Quantitative Mass Spectrometry
Quantification is used in most of our workflows as a solely qualitative protein analysis is not sufficient in most cases. The following image does show a classical scenario where quantitative MS is used for the comparison of different cell types or conditions (A), but also a use case where such a workflow is used to differentiate between bona-fide interactors and contaminats in an affinity-purfication MS experiment (B).
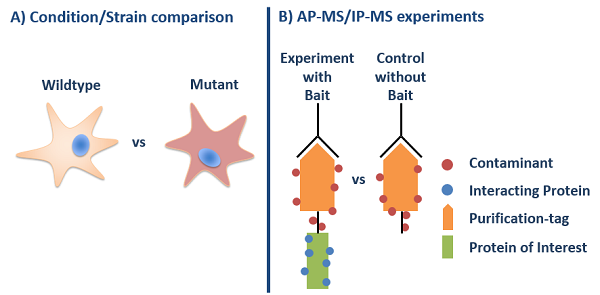
Quantification workflows
As intensity and concentration do not directly correlate to each other but are influenced by run-to-run differences and chemical properties of peptides, specific workflows are needed to perform accurate quantification by mass spectrometry.
We provide access to different quantitative methods: A) Metabolic Labelling like SILAC, B) Label-free quantification.
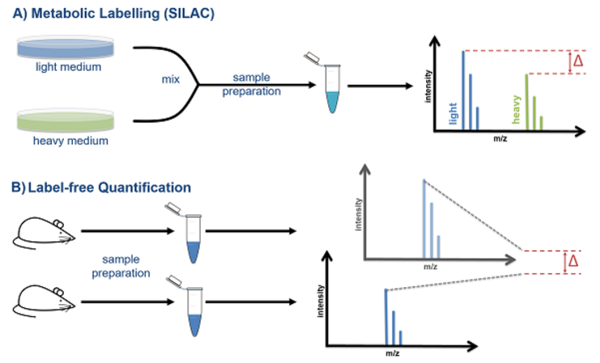
All methods have their inherent pros and cons and will be applied to match the specific requirements of an experiment.
