Precision Sarcoma Research
- Functional and Structural Genomics
- NCT
- Junior Research Group

Dr. Priya Chudasama
Group leader
We investigate molecular and clinical profiles of sarcomas through structural and functional genomics approaches to better understand sarcomagenesis and nominate biomarkers and precision therapeutics for clinical evaluation in sarcoma patients.

Our Research
Sarcomas are mainly divided into two types, bone sarcoma and soft-tissue sarcoma. Sarcomas display remarkable genetic and histologic diversity, as reflected by more than 150 subtypes according to the World Health Organization Classification, which in turn poses significant diagnostic and therapeutic difficulties. “Actionable” lesions that allow prediction of response to conventional or targeted anti-cancer drugs and/or represent direct targets for therapeutic intervention are lacking in the majority of cases due to incomplete understanding of the events that drive sarcoma development.
The Precision Sarcoma Research group funded by the Emmy Noether Program of the German Research Foundation (DFG) has been established with the aim to better understand the molecular alterations underlying tumor development and to identify novel targets for precision cancer therapy. To this end, we employ tumor multi-omics data generated within the in-house precision oncology workflows, in particular the NCT/German Cancer Consortium (DKTK) MASTER (Molecularly Aided Stratification for Tumor Eradication) as well as publicly available data resources to systematically investigate the genomic, epigenomic, transcriptomic and immunologic landscapes of sarcomas to identify pan-sarcoma or sub-entity specific pathognomonic alterations. In a project-specific manner, we have applied latest technology platforms at the DKFZ, such as long-read sequencing, single nuclei sequencing as well as spatial transcriptomics, to gain deeper insights into the biology of the tumors. Mechanistic investigations of selected aberrations are enabled by our expanding model system panel and comprehensive toolkit enabling functional genomics investigations (e.g. CRISPR/Cas9 libraries).
We are currently pursuing the following objectives:
- Targeting critical disease processes: Focus – Telomere maintenance
- Integrative multi-omics characterization: Focus – Ultra-rare sarcoma
- Development of innovative therapeutic approaches: Focus – Targeted protein degradation
These studies will improve the understanding of sarcomagenesis and identify targets for biological stratification and molecular mechanism-guided therapeutic intervention.
Strong support for ultra-rare sarcoma research
The Precision Sarcoma Research Group received a financial boost and solid encouragement from two very strong women – a loving mother and aunt of an ultra-rare sarcoma patient.
Mrs. Schwarz and Mrs. Fiebig-Frik are extremely talented in knitting, and love making Teddy bears and other decorative articles. In the cold but jolly Christmas market in Munich, they sold their latest creations, and, as a very touching gesture, donated all their proceedings to our research group. With the bottom of our hearts, we thank Mrs. Schwarz and Mrs. Fiebig-Frik for their generous contribution and recognition of our research.
This funding will support our research on ultra-rare sarcoma subtypes, which have the occurrence of ≤1 case per million people each year. Due to their extreme rarity, much less is known about the natural history, patient experience, underlying biology and effective strategies to diagnose and treat them. This is, in turn, due to insufficient patient samples and cancer models for lab-based research, and inherent difficulty in gathering patient populations for clinical trials. These limitations makes research on ultra-rare sarcomas particularly difficult—and often less eligible for traditional funding. Support such as received from Mrs. Schwarz and Mrs. Fiebig-Frik gives us this much-needed flexibility, and empowers us to learn from each tumor sample, each patient, and give access to this knowledge back to the scientific and patient community in a timely manner.
I am also grateful to Bastian Schwarz, a member of the DKFZ Patient Advisory council and a patient of ultra-rare sarcoma himself, with whom we are closely working on an initiative to bridge contact between patients to have access to latest knowledge, and researchers like us to have access to precious tumor samples. This partnership reflects the power of collaboration: patients, scientists, and donors united in the pursuit of knowledge and hope for those facing rare cancers. We will keep you updated.
Projects on Sarcoma

Telomeres are nucleotide sequences composed of 5’-TTAGGG-3’ tandem repeats that play an important role in maintaining genomic integrity by protecting the ends of chromosomes from DNA damage by means of the telomere-shelterin complex. Replication of telomeres is carried out by telomerase. To achieve replicative immortality, approximately 85% of cancers reactivate TERT expression. The remaining 15% cancers maintain telomere length via a telomerase-independent mechanism termed alternative lengthening of telomeres (ALT). Our previous work (Chudasama et al. Nat Comms 2018) and that of others has established ALT as a frequent feature in sarcomas that contributes to tumor progression. Being a cancer cell-specific process, ALT represents an attractive therapeutic target, however the landscape of ALT-targeting drugs is not well established. The interrogation of genes involved in ALT may reveal new biomarkers for targeted treatment of these aggressive tumors.
Thus far, we have screened >800 human sarcoma tumor samples to identify frequency of ALT in various sarcoma sub-entities. Following this stratification, we have employed integrative multi-omics analysis to identify genetic alterations that separate ALT-positive tumors from the ALT-negative tumors to identify ALT-associated aberrations that may be targetable directly or via secondary dependencies. Using a large panel of rare sarcoma cell lines and patient-derived xenografts, we are validating the “druggability” of candidate alterations and mechanistically characterizing those using phospho-proteomics to lay the groundwork for nominating ALT-specific targets for evaluation in the clinical setting. Moreover, we are developing methods to address the impact of heterogeneity in telomere maintenance mechanisms using machine learning tools (Belova et al. 2023 biorXiv) and spatially resolved technologies (Frank et al. Nucleic Acids Res 2022).
Limited availability of tumor samples, absence of model systems, lower number of patients for understanding of disease biology and drug development are critical hurdles in advancing basic, translational, and clinical research in rare cancers (<6 cases per 100,000/year) such as sarcoma. A subset of sarcoma, termed as ultra-rare sarcoma (≤1 case per 1,000,000, Stacchiotti S et al, 2021, Cancer, 27:2934) are exceedingly rare, with significant augmentation of above challenges. Our team aims to push forward ultra-rare sarcoma research by performing in-depth molecular characterization of tumor samples to inform diagnosis, classification, identification of pathognomonic alterations, model development, and entry points for biology-guided treatment.
Follicular Dendritic Cell Sarcoma
One example of ultra-rare sarcoma is follicular dendritic sarcoma (FDCS) that originates from follicular dendritic cells (FDCs). Diagnosis of FDCS is difficult due to histological similarities with many epithelial, mesenchymal, meningeal, or lymphoid malignancies. FDCS patients have poor prognosis and are still commonly treated with a lymphoma chemotherapy regimen (CHOP) despite evidence of mesenchymal origin of FDCs. We have assembled a cohort of >30 FDCS samples and have characterized the genomic and transcriptomic landscape of alterations, which led to identification of previously not described targetable lesions and oncogenic mechanisms, such as BRAFV600E mutations and telomerase overexpression by enhancer hijacking. Results of our analysis have led to clinical benefit to FDCS patient and we have identified a rare histological subtype of this ultra-rare sarcoma. We continue to characterize this intriguing disease using DNA methylation landscape and spatial technologies.
Alveolar soft-part sarcoma
Alveolar-soft part sarcoma (ASPS) is yet another ultra-rare sarcoma with poor prognosis in the metastatic setting. ASPS is driven by (X;17) (p11;q25) translocation to form the ASPSCR1-TFE3 fusion oncoprotein, with low number of additional genomic alterations, such as mutations or structural variants. We were intrigued by the finding of high level of immune cell infiltration in ASPS tumors, which according to the current dogma, is not a frequent feature of tumors of “silent” genomes. Furthermore, certain patients with ASPS tumors show exceptional response to checkpoint inhibition, which, however, does not correlate with expression levels of immune checkpoint molecules. We are employing transcriptome-based immune profiling approaches and spatial proteomics to 1) uncover the secrets of immune cell chemotaxis, 2) identification of a biomarker for response to checkpoint blockade treatment and 3) identify new mechanisms and vulnerabilities of fusion-driven modulation of immune microenvironment. To this end, in collaboration with group of Ana Banito (Soft-tissue Sarcoma, DKFZ and KiTZ) we have developed an immunocompetent ASPS mouse model that will allow pre-clinical validation of targets and biomarkers.
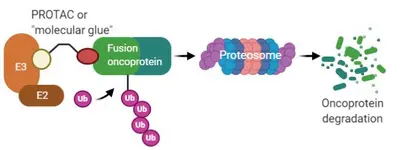
One third of sarcomas are characterized by recurrent genetic changes known as chromosomal translocations created by breakpoints within two cellular genes that result in generation of a chimeric fusion gene. In majority of cases, fusion genes in sarcoma involve chromatin remodeling factors or transcription factors. Gene fusion results in alteration in pattern of gene expression regulation, which results in – in majority of cases – enhanced proliferation, resistance to apoptosis, increased migration and invasion. As the fusion genes are the oncogenic drivers that are expressed only in tumor cells, they represent attractive molecular targets. However, fusion genes derived from transcription factors do not exhibit enzymatic activity and are hence not amenable to small-molecule inhibition. A promising approach towards that end is small molecules which induce proximity between E3 ubiquitin ligases and oncogenic substrates thereby targeting the substrates for degradation by the ubiquitin proteasome system (UPS). Success of this approach is exemplified by of compounds such as lenalidomide analogs, also known as immunomodulatory drugs (IMiDs) and proteolysis-targeting chimeras (PROTACs). By means of reporter-based functional genomic screens coupled with protein biochemistry and structural biology approaches, we are investigating protein degradation pathways of sarcoma fusion genes for nominating new E3 ligase-fusion oncoprotein pairs and structures for “molecular glue” or PROTAC design, pushing the development of new therapeutic for the “undruggable” sarcoma fusion oncoproteins.
Pan-Cancer Projects

Cancer metastases are strongest contributing factor to cancer-related mortality. Despite significant progress, the early detection and precise prediction of metastasis remains a major unresolved issue in cancer research. Funded by the National Decade Against Cancer Initiative of the German Federal Ministry of Education and Research, the innovative DECIPHER-M research project involves members from Aachen, Dresden, Essen, Heidelberg, Mainz and Munich who are pursuing an interdisciplinary approach that aims to decipher the complex mechanisms of cancer spread using state-of-the-art AI technologies.
DECIPHER-M addresses this with a unique approach using a new form of artificial intelligence (AI) called multimodal foundation models. In this project, these models are used to analyze a wide range of data, such as radiological images, pathological reports and genetic information of a patient together. This will answer fundamental questions about metastasis, such as the mechanisms of its occurrence, the potential to predict who might develop it and where, and what type of treatment might be most effective for different patients.
In addition, DECIPHER-M will provide practical tools that can be applied to individual patients to customize screening and treatment in cases at high risk of metastasis. Specifically, these tools aim to predict the most effective treatment for individual patients with metastatic disease so that these patients can be treated more effectively.
Priya Chudasama leads the Sub-project 8, which is dedicated to bridging foundational AI models with clinically actionable insights by elucidating the biological underpinnings of selected uni-modal and multi-modal signatures. With a strong research focus on sarcomas—a heterogeneous and rare group of malignancies—our team is eager to explore both their commonalities and distinctions relative to more common cancers, particularly in the context of mechanisms of metastasis development and diagnosis. This inquiry may help us tackle the challenge of data sparsity around rare cancers by implementing AI approaches. Finally, in collaboration with Cindy Körner and Markus Wartenberg of the NCT and DKFZ Patient Advisory Council as well as a nationwide network of patient representatives, Priya coordinates the patient-partnered research initiatives of the consortium.
Team
-

Dr. Priya Chudasama
Group leader
-

Wenxin Chao
MD student
-
Fenja Guyot
-
Sophie Herbst
-

Julien Picotto
Postdoctoral Researcher
Master/ MD thesis call
Selected Publications
Belova T, Biondi N, Hsieh PH, Chudasama P, Kuijjer M.
ALT-FISH quantifies alternative lengthening of telomeres activity by imaging of single-stranded repeats
Frank L, Rademacher A,…, Fröhling S, Chudasama P, Rippe K.
Small molecule-induced polymerization triggers degradation of BCL6
Stabicki M, Yoon H, Koeppel J, …, Chudasama P, …, Ebert B.
Integrative genomic and transcriptomic analysis of leiomyosarcoma
Chudasama P., Mughal S.S et.al
Get in touch with us