Neuroimmunology and Brain Tumor Immunology
- Immunology, Infection and Cancer
- Clinical Cooperation Unit

Prof. Dr. med. Michael Platten
Department Head
"We develop and bring to patients innovative personalized immunotherapies targeting brain tumors in a rigorous reverse translation approach."

Our Research
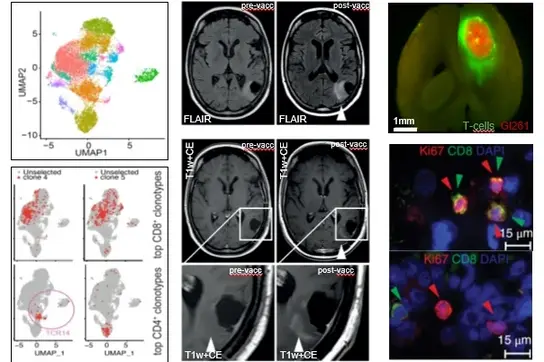
The central nervous system (CNS) is considered an immune privileged organ, where immune responses are tightly controlled through an intensive cross-talk with the peripheral immune system despite the blood-brain barrier. Our research group aims at developing and improving immune therapeutic approaches to target brain tumors by understanding molecular mechanisms of immunosuppression and exploiting novel immunotherapeutic treatment modalities. With expertise in comprehensive cellular and imaging-based analysis of tumor microenvironments, transcriptomics, and immunoreceptor profiling and utilization, and using clinical samples and mouse models, we strongly focus on clinical translation.
In the past years we have identified immunosuppressive functions and mechanisms of key metabolites that are produced by brain tumors. The discovery that TDO-derived tryptophan metabolites (kynurenines) drive growth of brain tumors and immunosuppression via the activation of the aryl hydrocarbon receptor (AHR) opened the way to novel therapeutic targets and implied further questions which we currently address using tumor models. A central goal is the identification of drugs interfering with tryptophan catabolism as potential therapeutics for malignant glioma. Our findings that IDH-mutant glioma cell-derived 2HG actively and directly inhibits adaptive cellular and innate immune responses by affecting immune cell function in the tumor microenvironment pave the way towards novel concepts of immunotherapeutic combination treatments which we are currently investigating in preclinical animal glioma models and chaperoning on their clinical translation.
The discovery of novel target antigens and T cell receptors for targeted immunotherapy of gliomas have been rigorously translated into first-in-human trials to show therapeutic efficacy of mutation-specific vaccine targeting clonal shared glioma driver mutations. Ongoing projects now focus on identification of further mutational antigens for specific immunotherapy and on specific TCR discovery for use in transgenic T cell therapy for glioma patients. Here, our main focus is on glioma-specific CD4+ T helper cells. In this regard, a workflow for the development of a patient-specific targeted immunotherapy for patients with gliomas based on mutanome analyses and T cell receptor identification have been developed. Bioinformatics-based approaches for tumor-infiltrating reactive T cell selection for such TCR discovery based on high throughput state-of-the-art single cell RNA, single cell VDJ, as well as TCRbeta deep sequencing, are boosting these processes and progress. We use synthetic immunology to engineer new T cell therapies for brain tumors.
Our Focus
We have developed long peptide vaccines targeting clonal shared driver mutations in gliomas such as IDH1.R132H or H3.K27M from preclinical validation in humanized mouse models to first-in-human clinical trials. These clinical trials have demonstrated that vaccine-induced mutation-specific CD4 T cells are capable of homing to the central nervous system and initiate a therapeutic inflammatory response. We put a strong emphasis on iterative optimization of vaccine responses by combining our vaccines with immune checkpoint inhibitors and testing them in "window-of-opportunity" trials allowing for reverse translation. High-throughput single cell sequencing of peripheral and tumor-infiltrating T cells allow for characterizing and validating glioma-specific T cell receptors on a patient-individual basis, which can be employed for T cell engineering.
To translate immunotherapeutic approaches from bench to bedside, we need to understand tumor-specific immune responses and study the full efficacy-spectrum from tumor regression to resistance. In humans this is complicated by many patient-individual factors, making careful study design and sensitive assays essential.
We are experts in human immune monitoring and work closely with our clinical partners to achieve deep-profiling of individual immune-responses. We have established a range of sensitive functional assays (ELISpot, ELISA, TIL expansion, epitope-specific expansion, flow cytometry) to be used across different types of patient samples (blood, CSF, tumor-tissue).
In parallel, we put particular emphasis on state-of-the-art sequencing-based technologies, such as bulk and single cell TCR and RNA sequencing, to understand the dynamics of T cell responses. This allows us to observe antigen- or treatment-induced changes in repertoire composition, and to trace and characterize individual T cell clonotypes across different samples/tissues and timepoints. TCR candidates with putative anti-tumor activity can be identified, recombinantly expressed and characterized in vitro with respect to specificity, affinity, avidity and HLA restriction. We use these receptors to fill a warehouse of tumor-reactive TCRs against antigens observed across patients, which could in the future be used as off-the-shelf therapeutics. To enable personalized TCR-based therapies, we are working to speed-up the discovery of receptors targeting patient-individual neoepitopes.
Personalised transgenic T cell therapies can – in some cases – cure a patient of cancer. However, until recently identifying patient specific, tumor-reactive T cell receptors took over 3 months. We are shortening this discovery phase to mere weeks by combining single cell sequencing, machine learning, and high-throughput, semi-automated cloning. We work closely with the T cell therapy engineering team to create new therapies for brain tumor patients.
Our machine learning model is available for academic use at https://predicTCR.com, for commercial use contact our spin-off https://www.tcelltech.eu/.
We are active in reverse translational studies, discovering tumor-reactive TCRs in samples from the lab’s ongoing clinical trials and testing them in preclinical models. We are working to develop such TCRs into off-the-shelf cell therapies for brain tumor patients, incorporating the latest technologies such as episomal DNA vector mediated TCR delivery in our upcoming INVENT4GB clinical trial. We also have longstanding interests in the role of HLA class II in brain tumors, the applications of immune monitoring CSF samples in rare disease, and understanding T cell trafficking into the brain
We aim to understand and tackle two of the main characteristics of immunologically cold brain tumors, which are (i) T cell exclusion due to growth behind the blood-brain-barrier in an immunologically special compartment, and (ii) a highly immunosuppressive microenvironment of the central nervous system, inhibiting cytotoxic T cells, both leading to unique resistance mechanisms. Comprehensive spatial, transcriptomic, and functional characterization of a heterogenous and complex interplay of T, myeloid, and stromal cells guides the way to novel combinatorial therapeutic concepts that are probed in various preclinical brain tumor models.
Research Teams
Immune monitoring and TCR discovery | German Cancer Research Center
MoreSelected Publications
Cancer Cell
News
Our MD student Kuralay Aman was awarded with the DAAD price for her social engagement for human rights and her brilliant academic performance.
Congratulation on this fantastic preformance!
MoreThe "Web of Science Group" of the US company Clarivate annually publishes a ranking of the world's most cited scientists in 20 different fields, covering all of science, medicine, economics and social sciences, as well as in the "Cross Fields" category. Twelve scientists who conduct research at the DKFZ or head joint bridging departments with the DKFZ made it to the very top in 2024: they are among the top one percent of the world's most cited researchers in their respective fields. Scientists whose work is cited particularly frequently by peers are considered to have above-average recognition in their field. The frequency of citations is therefore one of the most important measures of the influence and performance of individual researchers.
Michael Platten is one of ten winners in the Life Sciences category and ultimately the winner of the Breakthrough of the Year 2024 in the Life Sciences. Congratulations!!
The title "Falling Walls Science Breakthrough of the Year" is awarded annually. It recognizes the most significant scientific breakthroughs in various categories of the Falling Walls Global Call. The Falling Walls Science Summit focused on topics such as artificial intelligence, renewable energies, quantum research and science diplomacy. The Berlin-based Falling Walls Foundation aims to bring together people who want to "break down the next walls in science and society". Since the first conference on the 20th anniversary of the fall of the Berlin Wall in 2009, Falling Walls has developed into a constantly growing network of the most ambitious and forward-thinking minds from around the world.
Press release: Life Sciences | Falling Walls
Prof. Michael Platten took over the presidency of the European Association of Neuro-Oncology (EANO) at this year's congress in Glasgow. With over 700 members from more than 70 countries, it is one of the most important neuro-oncology societies in the world. Michael Platten will hold this honorable office for two years.
More news can be found in our archive: German Cancer Research Center: News Archive
Staff
-

Prof. Dr. med. Michael Platten
Department Head
-

Belize Acharya
MD-Student
-

Dennis Alexander Agardy
PostDoc
-

Kuralay Aman
MD Student
-
Lucas Arnoldt
-

Isabelle Bernhardt
MD Student
-
Jens Blobner
-

Dr. Tamara Boschert
PostDoc
-
Berin Boztepe
-

Dr. Theresa Bunse
Teamleader
-

Dr. Lukas Bunse
Teamleader
-

Yu-Chan Chih
PostDoc
-

Amelie Christina Dietsch
MD Student
-

Andreas Dobbelstein
MD Student
-

Alexander Ernst
Technical Assistance IMU
-

Alina Errerd
Master Student
-

Henrike Feldmann
PhD Student
-
Lena Fleischhacker
-

Hannah Gelhaus
PhD Student
-

Julia Gellert
MD Student
-
Dr. Isabel Göhring
-

Niklas Graßl
PostDoc
-

Dr. Edward Green
Teamleader
-
Moritz Hamberger
Master Student
-

Julian Hlawatsch
MD Student
-
Melissa Höpfner
Trainee
-
Zeren Hu
-

Ingrid Hülsmeyer
Technical Assistance IMU
-

Kristine Jähne
Technical Assistance
-
Gyuyeon Jang
-

Simone Jünger
Technical Assistance IMU
-

Dr. Thomas Kerzel-Gnaneswaran
-

Vahid Khaki Bakhtiarvand
PhD Student
-
Claudia Koch
-

Philipp Koopmann
MD Student
-

Tom Kuhn
PostDoc
-

Dr. Katharina Lindner
PostDoc
-

Claudia Maldonado Torres
Technical Assistance
-
Dr. Iris Mildenberger
-

Sarwar Mustafa
MD Student
-

Marie-Therese Neuhoff
PhD Student
-

David Palmero Canton
PhD Student
-

Alina Paul
PhD Student
-

Dr. Isabel Poschke
Teamleader
-

Jakob Rosenbauer
PostDoc
-

Samira Moreen Sachse
Technical Assistance IMU
-

Dr. Katharina Sahm
Teamleader
-

Georgios Samaras
PhD Student
-

Khwab Sanghvi
PostDoc
-
Sara Santo
-
André Maximilian Scheiner
-

Robi Schramm
MD Student
-
Robin Seitz
Technical Assistance
-

Akanksha Shukla
Technical Assistance IMU
-

Ina Simeth
-

Saskia Stange
PhD Student
-

Dr. Chin Leng Tan
PostDoc
-

Clara Tejido Dierssen
PhD Student
-

Verena Turco
-
Martin Uerlich
-
Lilith Hira Ükil
-

David Vonhören
PhD Student
-

Dr. Marie-Christine Wagner
Lab and project management
-

Binghao Zhao
MD Student
Get in touch with us














