Synthetic Biology
- Copying Life: synthesis of an enzymatically active mirror-image DNA-ligase made of D-amino acids
- Establishment of a fully synthetic mirror-image biological system (MirrorBio)
- Exploiting sequence and stability information for directing nanobody stability engineering
- Structural Modelling: Revised CHARMM force field parameters for iron-containing co-factors of photosystem II
Synthetic Biology and Systems Biology have become key research areas in the quest for understanding living cells. What is missing is an experimental system in which the knowledge from system-wide analyses could be artificially reproduced and studied. Our goal is to create a mirror-image synthetic biology: that is, to mimic, entirely independent of nature, a biological system and to re-create it from artificial component parts.
Starting from enantiomeric L-nucleotides and D-amino acids, we intend to use chemical synthesis to establish a functioning mirror-image biological system. Several basic DNA-DNA, DNA-protein and protein-protein interactions as well as the functions of few enzymes will be copied from nature into a totally synthetic artificial system based on L-DNA and D-proteins. This will require a few D-form enzymes and L-DNA sequences plus co-factors that would form a basic enantiomeric system. In addition, binder molecules, such as nanobodies and DARPins, and transcription factors, or the DNA-binding domains thereof, are generated and studied in comparison to their natural partners. We actively pursue the production of molecules, which exhibit superior performance, by also modulating parameters such as stability and aggregation behaviour. This is complemented by modelling so as to define and predict structural feature that are critical for molecules' activity. Next to the establishment of components for artificial biology, there are immediate practical utilities that are aimed at within this project, such as the protection of therapeutic binders from degradation as they will not be substrates of natural proteases, permitting oral application, for example.
In the long run, the objective is to set up a self-replicating system, eventually including D-protein production. While this is still some way off, it would offer a major advance in medicine. Molecules produced in such a system could overcome several of the limitations that are currently still inherent to therapeutic proteins.
Copying Life: synthesis of an enzymatically active mirror-image DNA-ligase made of D-amino acids
We report the synthesis of a functional DNA-ligase in the D-enantiomeric conformation, which is an exact mirror-image of the natural enzyme, exhibiting DNA ligation activity on chirally inverted nucleic acids in L-conformation, but not acting on natural substrates and with natural co-factors. Starting from the known structure of the Paramecium bursaria chlorella virus 1 DNA-ligase and the homologous but shorter DNA-ligase of Haemophilus influenza, we designed and synthesized chemically peptides, which could then be assembled into a full-length molecule yielding a functional protein. The structure and the activity of the mirror-image ligase were characterized, documenting its enantiospecific functionality.
Publications:
Weidmann et al. (2019) CELL Chem. Biol., in press. [PDF]
Establishment of a fully synthetic mirror-image biological system (MirrorBio)


The basic motivation behind the project is the ambition to understand biological processes to an extent that will permit their re-creation. This ability would be documented best, if functioning molecular systems could be established that are entirely independent of any natural compounds but generated by initially only chemical processes for the production of synthetic biomolecules and their assembly into artificial molecular systems. Also, artificial experimental systems will complement current Systems Biology, evaluating biological models experimentally. Similar to what is ongoing in physics, insight into cellular functioning will be gained by an iterative processing of information resulting from experimental and theoretical analyses. Eventually, this may lead to an archetypical model of a cell.
The project is performed as an EraSynBio consortium with partners from three other institutions:
- Philip E. Dawson, The Scripps Research Institute, USA
- Andreas Plückthun, University of Zürich, Switzerland
- Jussi Taipale, University of Helsinki, Finland
Publications:
Weidmann et al. (2016) Org. Lett. 18, 164-167. [PDF]
Olea et al. (2015) Chem. Biol. 22, 1437-1441. [PDF]
Hauser et al. (2006) Nucleic Acids Res. 34, 5101-5111. [PDF]
Exploiting sequence and stability information for directing nanobody stability engineering
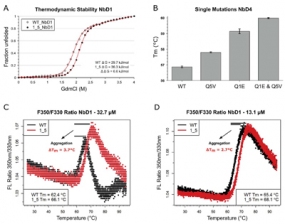
Mechanism of nanobody stabilisation by N-terminal mutations (Q1E and Q5V). (A) The thermodynamic stability of nanobody NbD1 and its N-terminally mutated variant was measured in guanidinium chloride dependent equilibrium unfolding experiments. Unfolded protein was measured by intrinsic tryptophan fluorescence. Red lines represent fitted curves. (B) Additive effect of thermostabilisation by single and double mutations in nanobody NbD4, measured by differential scanning fluorimetry in triplicate. Improvements by mutations Q5V (0.9 °C) and Q1E (2.3 °C) match the stabilisation in the double mutant (3.1 °C). (C, D) Tryptophan fluorescence ratio (350 nm/330 nm) for melting nanobody NbD1 and its N-terminally mutated variant; in panel C, a concentration of 32.7 μM was used; in panel D, concentration was reduced to 13.1 μM. Aggregation is indicated by a reduced amplitude of the unfolding transitions in the fluorescence traces and can be quantified by comparing Tm values of both concentration sets. Heating rate: 0.5 °C/min.
© dkfz.de
Variable domains of camelid heavy-chain antibodies, commonly named nanobodies, have high biotechnological potential. In view of their broad range of applications in research, diagnostics and therapy, engineering their stability is of particular interest. Towards these ends, we analyzed the sequences and thermostabilities of 78 purified nanobody binders. From this data, potentially stabilizing amino acid variations were identified and studied experimentally. Some improved the stability of nanobodies by up to 6.1°C, with an average of 2.3°C across eight modified nanobodies. The stabilizing mechanism involves an improvement of both conformational stability and aggregation behavior, explaining the different effect on individual molecules. Other potentially stabilizing variations actually led to thermal destabilization of the proteins. The reasons for this contradiction between prediction and experiment were investigated. The results illustrate the potential and limitations of engineering nanobody thermostability from a medium-throughput data set and indicate a species-specificity of nanobody architecture.
Publications:
Kunz et al. (2018) Sci. Rep. 8, 7934. [PDF]
Kunz et al. (2015) BBA-Gen. Subjects 1861, 2196-2205. [PDF]
Structural Modelling: Revised CHARMM force field parameters for iron-containing co-factors of photosystem II

Cover image of the Journal of Computational Chemistry, volume 39, issue 1, on 5 January 2018. It presents artwork that is based on the results reported in the above mentioned publication.
© dkfz.de
Photosystem II is a complex protein–cofactor machinery that splits water molecules into molecular oxygen, protons, and electrons. All-atom molecular dynamics simulations have the potential to contribute to our general understanding of how photosystem II works. To perform reliable all-atom simulations, we need accurate force field parameters for the cofactor molecules. We present here CHARMM bonded and non-bonded parameters for the iron-containing cofactors of photosystem II that include a six-coordinated heme moiety coordinated by two histidine groups, and a non-heme iron complex coordinated by bicarbonate and four histidines. The force field parameters presented here give water interaction energies and geometries in good agreement with the quantum mechanical target data.
Publications:
Adam et al. (2018) J. Comput. Chem. 39, 7-20. [PDF]