RNA-Protein Complexes and Cell Proliferation
- Functional and Structural Genomics

Priv. Doz. Dr. Maiwen Caudron-Herger
Once per cell cycle, cells face the challenging task of accurately segregating their chromosomes into two identical sets. Although cell division is highly regulated, errors may occur, leading to either cell death or the genome instability that underlies many human diseases such as cancer. RNA-protein complexes are critical regulatory elements in important cellular processes, including cell division, which we are investigating.
Image: © MCH

Image: © MCH
“Cells have the wonderful ability to assemble a great variety of dynamic structures performing very complex and specific tasks. Our goal is to improve our understanding of the underlying regulatory mechanisms and to answer the question of how dysfunction can lead to disease.”
PD Dr. Maïwen Caudron-Herger
Our Research
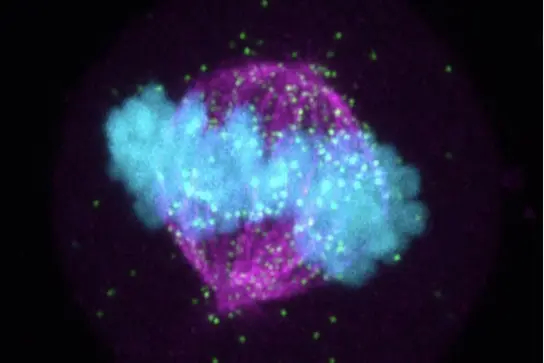
Despite significant advances in cell biology, the complex processes that regulate and drive cell division are still not fully understood, creating opportunities for discoveries related to the causes and vulnerabilities of highly proliferative diseases such as cancer.
RNA-protein complexes have emerged as critical regulatory elements in multiple key cellular processes with significant relevance to health and disease, making them attractive targets for research. Our current projects address the pivotal role of RNA and RNA-protein complexes in orchestrating the functional assembly of critical mitotic structures for the error-free completion of cell division. By integrating the RNA component, our approach adds a new level of complexity to the analysis of key events in this fundamental phase of the cell cycle.
Our approach is characterized by its interdisciplinary and collaborative nature, encompassing a diverse spectrum of methodologies. These include immunoprecipitation to probe protein-protein interactions, UV crosslinking and immunoprecipitation (iCLIP)-based approaches to investigate protein-RNA interactions, high-throughput sequencing and bioinformatic analyses to identify interacting RNA transcripts, and confocal microscopy imaging to localize proteins, RNAs and their interactions.
In addition to the impact on our basic understanding of cell division, the newly acquired knowledge has great potential to be translated into innovative cancer therapy strategies by targeting yet unexplored cellular functions associated with specific RNA-protein interactions.
Important Concepts
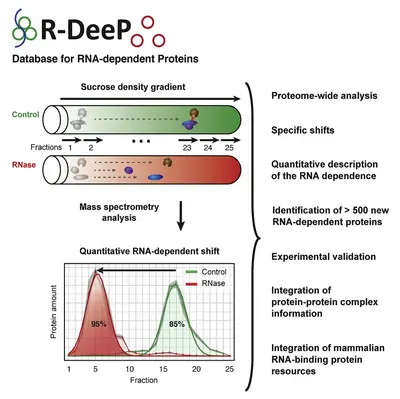
We recently developed the concept of RNA dependence, where we defined a protein as RNA dependent when its interactome depends on the presence of RNA. This concept has been translated into a comprehensive proteome-wide, unbiased and quantitative screening method called R-DeeP.
R-DeeP is based on the fractionation of cellular lysate – with and without prior RNase treatment – by sucrose density gradient ultracentrifugation and subsequent analysis by proteome-wide mass spectrometry or Western blotting of individual proteins.
R-DeeP determines the ability of a protein to form protein complexes exclusively in the presence of RNA by direct or indirect interaction with the RNA molecules. In the R-DeeP screen, we identified a significant enrichment for microtubule-related proteins. This indicates previously unknown potential functional implications of RNA-protein interactions, particularly in the context of cell division.
Check our updated database at R-DeeP3.dkfz.de.
Related publications:
- Rajagopal et al. 2025 - Nature Communication. 16:2325
- Caudron-Herger et al. 2020 - Nature Protocols. 15:1338-1370
- Caudron-Herger et al. 2019 - Mol Cell. 75:184-199
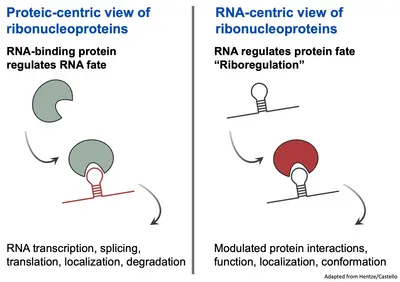
Our understanding of RNA-protein interactions has been predominantly protein centric, emphasizing the role of proteins as active agents controlling the fate of RNA throughout its lifecycle.
However, the massive effort to comprehensively identify the proteins in the cells interacting with RNA has lead to an ever-growing catalogue of RNA-binding proteins (RBPs) in several species. Surprisingly, the RBPs encompasse a majority of proteins lacking canonical RNA-binding proteins, refered to as “unconventional” RBPs. These unconventional RBPs are proteins previously known for their roles in various biological processes often unrelated to RNA and their interactions with RNA had been so far overlooked.
The exploration of the relationship between unconventional RBPs and RNA has led to an exciting concept termed ‘riboregulation’. Riboregulation challenges the classical view of RBP controlling RNA. Instead, it proposes that RNA itself actively regulates protein localization, conformation, interactions and functions. Thus, this novel paradigm highlights RNA not just as a passive substrate but as an active regulator of the interacting proteins.
Related publications:
Our Projects
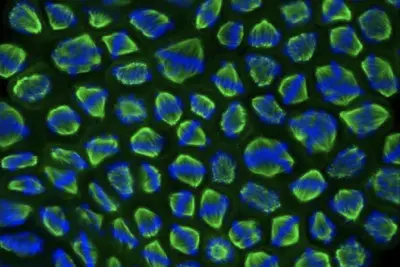
RNA-dependent proteins during cell division
We have created an atlas of RNA-dependent proteins (see concept above) which highlights a great number of mitotic factors as unconventional RNA-binding proteins: they seem to bind to RNA in absence of known RNA-binding domains. This suggests an important role for RNA during cell division in multiple parts of the cell division machinery (microtubules, condensed chromosomes, kinetochores, centrosomes etc ...), which we aim to understand. In particular, our projects investigate the role of mitotic unconventional proteins in the context of breast and ovarian cancers using a multidisciplinary approach.
Funding and support:
- Dr. Rolf M. Schwiete Stiftung
- Health + Life Science Alliance Heidelberg Mannheim
- Baden-Württemberg Stiftung
Related publications:

The characterisation of RNA-binding proteins
The analysis of RNA-protein complexes is central to elucidating the complex molecular networks that govern cellular processes. With the increasing recognition of their importance, proteins that interact directly with RNA have attracted growing interest. However, the comprehensive but specific identification of such RNA-binding proteins (RBPs) and the dissection of their functions in association with RNA remain major challenges in the field of RNA biology.
To further deepen our knowledge of RBPs, several proteome-wide strategies have been developed to identify RBPs in different species. Thus, a large number of studies have provided catalogs of experimentally identified as well as predicted RBP candidates. The rapid evolution of the field has led to an accumulation of isolated datasets, hampering both accessibility and comparability. Furthermore, tools for linking RBPs to cellular pathways and functions or diseases were lacking. Driven by our own research and as a service to the scientific community, we developed RBP2GO, a comprehensive database of all available proteome-wide datasets for RBPs across 13 species, providing a catalog of 29215 RBP candidates (RBP2GOv2.DKFZ.de). It not only includes data on RBP interaction partners but also provides information on related biological processes, molecular functions, and cellular components, as well as relation to diseases via the Disease Ontology (disease-ontology.org) and the DISEASES database (diseases.jensenlab.org).
RBP2GO provides a user-friendly web interface with a scoring system that reflects the potential of RBP candidates to be bona fide RBPs. The RBP2GO advanced search provides users with options to navigate through the interconnected datasets to promote and stimulate new research directions.
Ongoing projects are currently refining the list of RBPs by incorporating information on RNA-binding domains (RBDs) and RNA-related protein families. These multi-purpose analyses aim to refine the extensive list of RBP candidates and to provide a list of high confidence RBPs, based on a scoring system.
Related publications:
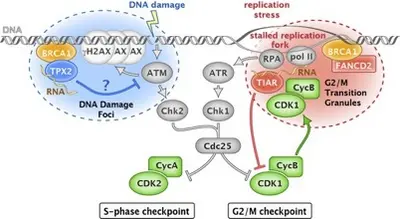
RNA-binding proteins in breast and ovarian cancers (RB2OC)
Cancer is a complex disease that involves the successive establishment of dysfunctions in multiple key cellular processes leading to characteristics hallmarks such as sustaining proliferative signaling, resisting cell death and enabling replicative immortality. While breast cancer is the most prevalent cancer in women, ovarian cancer has one of the poorest prognoses among all cancer types. Mutations in BRCA1 and BRCA2 represent major risk factors for both breast and ovarian cancer. While hereditary breast and ovarian cancer represent ∼10%–20% of the disease fraction in the population, only ∼10% of hereditary cases are due to mutations in BRCA1/2, suggesting additional, yet unknown risk factors to be identified.
RNA-binding proteins play important roles in the regulation of gene expression through the regulation of mRNA splicing, stability and translation, thereby affecting the development, progression and treatment response of cancer diseases. Recent advances in RNA biology have led to the identification of an increasing number of RNA-binding proteins. Based on our data, RNA-binding proteins such as TIAR and TPX2 are important for cell cycle control and genomic stability, and connected to BRCA proteins. RB2OC will explore their respective role and relationship in both cancer types. In particular, we will examine the potential diagnostic and prognostic value of TIAR and TPX2 expression levels, assess their relation to DNA repair pathways through mutational landscape analysis, and address the impact of TIAR/TPX2 on the sensitivity of breast cancer and ovarian cancer to specific DNA damage inhibitors. Using an interdisciplinary approach, we will determine the role of TIAR and TPX2 dysfunction as putative drivers of breast cancer and ovarian cancer pathogenesis.
Collaboration:
Funding:
Our Team
-

Priv. Doz. Dr. Maiwen Caudron-Herger
Group Leader
-

Dr. Simona Cantarella
Postdoc
-

Jens Feugmann
PhD Student
-

Jeanette Simone Seiler
Technical Assistant
-

Jana Theiß
Technical Assistant
-
Nina Bank
Student, Master Molecular Biotechnology
Selected Publications
Simona Cantarella, Jule Neffe, Malte Hermes, Fabio Rauscher, Robert Schwarz, Praarthanaa Jaisankar, Enno Schäfer, Maïwen Caudron-Herger
Varshni Rajagopal, Jeanette Seiler, Isha Nasa, Simona Cantarella, Jana Theiß, Franziska Herget, Bianca Kaifer, Melina Klostermann, Rainer Will, Martin Schneider, Dominic Helm, Julian König, Kathi Zarnack, Sven Diederichs, Arminja N Kettenbach and Maïwen Caudron-Herger
Wassmer E, Koppány G, Hermes M, Diederichs S and Caudron-Herger M.
Caudron-Herger M*, Ralf E. Jansen, Elsa Wassmer and Sven Diederichs. *Corresponding author.
Caudron-Herger M*, Barreau E, Nasa I, Schultz A, Seiler J, Kettenbach AN and Diederichs S*. *Corresponding authors.
Caudron-Herger M*, Rusin SF, Adamo ME, Seiler J, Schmid V, Barreau E, Kettenbach AN and Diederichs S*. *Corresponding authors.
Our Databases

RBP2GO provides a comprehensive database of RNA-binding or RNA-dependent proteins from all available proteome-wide studies in 13 different species. It includes the annotation of their functions as well as interaction partners - and allows also reverse searches for RNA-binding proteins with specific molecular functions, biological processes, cellular compartments or a known link to diseases. RBP2GO is published in Cantarella et al, Nucleic Acids Research, 2025.

R-DeeP3 provides the proteome-wide screening results for RNA-dependent protein complexes from unsynchronized HeLa S3 and A549 lung cancer cells as well as synchronized HeLa cells in mitosis and interphase. In addition, R-DeeP3 offers options for the comparison of all the datasets and for the convenient download of the results.
Teaching at Heidelberg University
Faculty of Engineering Sciences
- Methoden der Bioinformatik: Datenanalyse und Bildanalyse (BSc Molecular Biotechnology, winter semester)
- Anwendung bioinformatischer Systeme: Data Analysis (BSc Molecular Biotechnology, summer semester)
- Frontiers in Biosciences: Molecular, Genetic and Bioinformatic Approaches in Cancer Research (Major Cancer Biology, winter semester)
- Physical Methods in Systems Biology (Major Systems Biology, summer semester)
Additional Seminar
- How to design a scienctifc presentation - Visual communication for scientists

Get in touch with us

