Pediatric Glioma Research
- Functional and Structural Genomics
- KiTZ

Prof. Dr. David Jones
Division Head
Gliomas are the most common form of brain tumor found in children, in total accounting for about half of all central nervous system tumors in 0-19 year olds. Two major categories are usually distinguished: low-grade and high-grade glioma.
Our Research
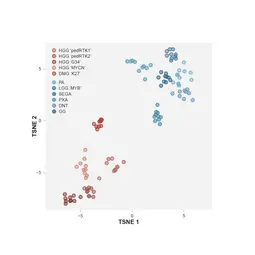
Low-grade gliomas are a histologically and biologically varied collection of tumor entities, which are often defined by genetic alterations in the mitogen-activated protein kinase (MAPK) pathway and are typically considered as benign tumors due to their good overall survival rates. This survival, however, often comes at a significant cost to quality of life due to effects of treatment as well as the tumor itself. Multiple recurrences over several years are also not uncommon, creating a heavy burden on patients and their families. Our research aims to learn more about the background of these tumors and their unusual growth behaviour, as well as to identify treatments that are more tailored to individual tumor biology and therefore hopefully have fewer side effects.
High-grade gliomas, in contrast, carry an extremely dismal prognosis. These highly aggressive tumors, most commonly glioblastoma or diffuse intrinsic pontine glioma (DIPG), typically display a much greater degree of genomic instability than lower-grade lesions. Combined disruption of multiple cellular processes leads to rapidly dividing cancer cells that diffusely infiltrate the surrounding normal brain tissue, with an almost universally fatal outcome. Our research in this area aims at understanding the heterogeneity both within single tumors and between individuals, to identify the most important patterns of alterations in the cellular machinery. We then try to recapitulate these changes in model systems, in order to understand their contribution to a cell becoming cancerous and also to test new therapies in a rationally targeted manner.
All of the work being conducted in the lab is based on the application of cutting-edge genomic (next-generation DNA/RNA sequencing), epigenomic (DNA methylation, ChIPseq), and functional technologies (CRISPR/Cas9, somatic gene transfer models) to enhance our understanding of the biological underpinnings of pediatric brain tumors. A further important focus of the group is the translation of this research into practical applications for the benefit of patients. To this end, we are closely involved in the coordination of two international molecular diagnostic programs: the INFORM study for identification of possible drug targets in high-risk pediatric cancers; and the MNP2 study to investigate the value of molecular analysis as a tool for accurate classification of brain tumors.
More information on the Pediatric Neurooncology community in Heidelberg can be found here.
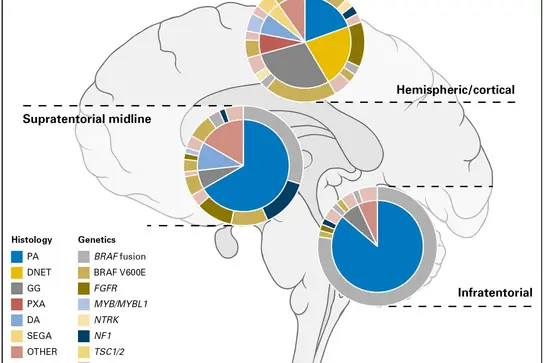
Approximation of the frequencies of different histological groupings of pediatric low-grade glioma and associated molecular genetic alterations, split by location within the brain. Adapted from Sturm D, Pfister SM and Jones DTW, J Clin Oncol 2017.
Projects
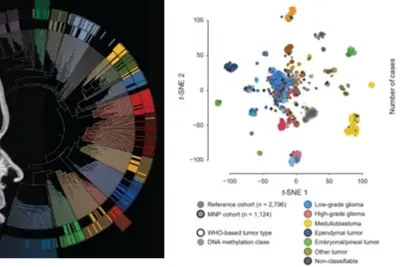
Gliomas are the most common type of central nervous system (CNS) tumor in children and adolescents. Epigenetic analysis has become an indispensable tool in pediatric brain tumor research, supporting diagnosis and revealing the molecular mechanisms driving tumor development. DNA methylation signatures, combined with histopathological and molecular tumor classification, are globally recognized and have been integrated into the latest WHO guidelines for the classification of CNS tumors. Our research focuses on epigenetic modifications, including DNA methylation patterns and histone modifications, which influence gene expression. By identifying specific epigenetic changes in pediatric gliomas, we aim to develop improved diagnostic tools and targeted therapies. Our results have already provided valuable insights into the biology of these tumors and are promising for the development of more effective treatments in the future.
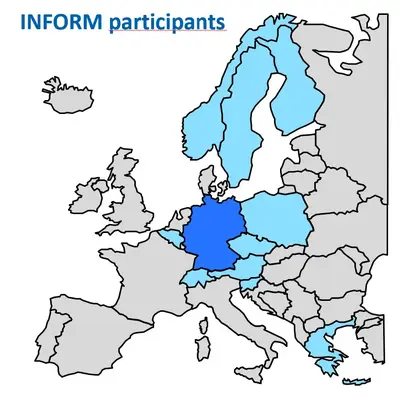
We play a central role in organizing the international precision oncology program for relapsed malignancies in children (INFORM) by overseeing molecular analysis and real-time data interpretation. This provides access to comprehensive molecular profiles and rapid drug screening for high-risk pediatric cancer patients across Europe. The INFORM network spans 13 European countries, is active in over 100 pediatric oncology research centers and has enrolled more than 3,300 relapsed or refractory pediatric cancer patients, almost half of whom come from international partners. The first clinical response analysis showed a survival benefit for patients with high-priority targets who received appropriate treatments. The program has also successfully integrated a platform for creating personalized ex vivo drug sensitivity profiles in real-world settings and provides functional data for clinical decision making. Significant strides have been made in improving the logistics, methodology, and efficacy of personalized diagnosis and treatment. These include the transition to high-resolution whole-genome sequencing (WGS) as a new standard for molecular diagnostics. Importantly, this routine analysis is reimbursed by German health insurance providers and is helping to establish similar sequencing and screening platforms in other countries.

Low-grade brain tumors are the most common type of CNS tumor in children, accounting for about half of all brain tumors in children. In Germany and the UK, over 400 children and adolescents are diagnosed each year. These tumors arise from DNA alterations that cause uncontrolled cell growth in the brain or spinal cord. Although they grow slowly and are unlikely to spread, their treatment is challenging due to their location and often results in long-term health issues.
The Everest Center for Low-Grade Pediatric Brain Tumors – a collaboration between research groups in Berlin, Jena, London and Heidelberg (KiTZ & DKFZ) – is dedicated to advancing understanding and developing new treatments. Established in 2017, the center is funded by The Brain Tumor Charity and has raised 12 million euros (10 million pounds) thanks to the inspiring Everest in the Alps fundraising campaign.
A close partnership with Boston's Dana-Farber Cancer Institute strengthens the research effort and ensures that discoveries reach young patients. In addition, the project focuses on improving quality of life by tracking individual patients' experiences over time using advanced technologies.
The Everest Center's name honors an extraordinary fundraising effort inspired by Toby, a young patient. Since 2015, a dedicated team of skiers has crossed the Alps, overcoming a vertical equivalent to Mount Everest (8,848m), and raised over £10 million for research. Their commitment is invaluable – on behalf of all affected families, we would like to express our deepest gratitude to them!

The Dietmar Hopp Foundation is supporting to set up an international clinical "data depot" at the Hopp Children's Cancer Center Heidelberg (KiTZ) under the direction of David Jones and Stefan Pfister. This project aims to create the basis for children and adolescents to benefit quickly from cancer drugs already approved for adults.
By collecting and integrating clinical data from various international sources, the "data repository" aims to accelerate access to more effective therapies with fewer side effects for young cancer patients. This will enable existing drugs to be used more quickly for the treatment of children and adolescents without having to wait for lengthy development processes for new drugs.
In addition to the "data depot", the research and development of innovative treatment methods is also supported in order to improve the chances of recovery and quality of life for young patients.

MNP Int-R is a continuation and follow-up study of MNP 2.0, which established the use of DNA methylation signatures as part of an integrated histological and molecular classification to improve the accuracy of pediatric brain tumor diagnoses at initial diagnosis. This initiative paved the way for the incorporation of molecular diagnostics, including DNA methylation profiling and targeted gene panel sequencing, into routine diagnostics for primary CNS tumors in children and adolescents. Integrated reference analysis is helping to identify previously unknown tumor types and subtypes, reveal their biological relationships, and classify rare molecularly defined tumor entities. Further refinement and training of the DNA methylation-based classification system improves molecular diagnostics and shapes the design of future therapeutic trials, advancing personalized precision treatments and novel targeted therapy approaches. By integrating pathological, radiological and molecular data into a comprehensive tumor diagnosis, MNP Int-R aims to maintain high diagnostic standards in an international setting and to significantly influence clinical disease management. Furthermore, ongoing data collection will further refine diagnostic accuracy and treatment strategies.
Our Team
Group Picture
Publications
2019 - 2025
MoreErnst, K.
Establishment and evaluation of single-cell RNA-sequencing methods to investigate tumor heterogeneity in newly generated pediatric glioma models
Deng, M. Y.
Molecular identification and characterization of novel pediatric brain tumor entities through genome-wide DNA methylation profiling
Sommerkamp, A.
Molecular comparison, preclinical modeling and improved diagnostics of pediatric low-grade gliomas
Ismer, B.
Novel gene fusions identified as new drug targets in paediatric glioma and their pre-clinical characterization
Maile, J.
Profiling of epigenetic-related vulnerabilities in K27M diffuse midline glioma using CRISPR/Cas9 genetic screening
Selected Publications
Sturm D, Andreiuolo F, Gessi M, Kölsche C, …, Pietsch T*, Sahm F*, Pfister SM*, Jones DTW*
FOXR2 is an epigenetically regulated pan-cancer oncogene that activates ETS transcriptional circuits
Tsai JW, Cejas P, Wang DK, Patel S, …, Long H, Jones DTW*, Bandopadhayay P*, Phoenix TN*
Deng MY, Sturm D, Pfaff E, Sill M, …, Korshunov A, von Deimling A, Pfister SM, Jones DTW
Clarke M, Mackay A, Ismer B, Pickles JC, …, Ellison DW*, Jacques TS*, Jones DTW*, Jones C*
Jones DTW, Banito A, Grünewald TGP, …, Schleiermacher G, Smith MA, Westermann F, Pfister SM
Capper D*, Jones DTW*, Sill M*, Hovestadt V*, …, Brandner S, Korshunov A, von Deimling A, Pfister SM
Gröbner SN, Worst BC, Weischenfeldt J, Buchhalter I, …, Jones DTW, Lichter P, Chavez L, Zapatka M, Pfister SM
Mackay A, Burford A, Carvalho D, Izquierdo E, …, Jones DTW, Fouladi M, von Bueren AO, Baudis M, Resnick A, Jones C
Bender S, Gronych J, Warnatz H-J, Hutter B, …, Yaspo ML, Pfister SM*, Lichter P*, Jones DTW*
Worst BC, van Tilburg CM, Balasubramanian GP, Fiesel P, …, Capper D, Pfister SM*, Jones DTW* and Witt O*
Bender S, Tang Y, Lindroth AM, Hovestadt V, Jones DTW, …., Monje M, Plass C, Cho YJ, Pfister SM
Alexandrov LB, Nik-Zainal S, Wedge DC, Aparicio SA, …, Shibata T, Pfister SM, Campbell PJ, Stratton MR
Jones DTW*, Hutter B*, Jäger N*, Korshunov A, …, Jabado N, Eils R, Lichter P and Pfister SM
Schwartzentruber J*, Korshunov A*, Liu XY*, Jones DTW, …, Plass C, Majewski J, Pfister SM, Jabado N
Jones DT, Kocialkowski S, Liu L, Pearson DM, Bäcklund LM, Ichimura K, Collins VP
Get in touch with us



