Structural Biology of Infection and Immunity
- Immunology, Infection and Cancer

Dr. Erec Stebbins
Head of Division/Abteilungsleiter
We are mechanistically characterizing the antigenically distinct variant surface glycoproteins (VSGs) of the African trypanosome and their interaction with antibodies, employing a model system to understand long-term immune evasion, a process underlying malignancies.
Image: Left: Images of the African trypanosome replicating in the blood, its surface coat shown at the bottom as it switches from one VSG to the other. Right top: protein crystal and diffraction pattern. Right bottom: Structure of VSGsur bound to the anti-trypanosome drug suramin.,
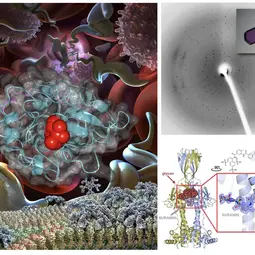
Image: Left: Images of the African trypanosome replicating in the blood, its surface coat shown at the bottom as it switches from one VSG to the other. Right top: protein crystal and diffraction pattern. Right bottom: Structure of VSGsur bound to the anti-trypanosome drug suramin.,
Our Research
Immune escape variants typify long-term pathogenesis in both infectious disease and malignancy. The immune system recognizes foreign and dangerous "self" antigens, clearing cells harbouring them such as microbes and tumours. Pathogens and persistent cancers often counter immunity by varying surface antigens. We are using a controlled, bio-safe, genetically tractable model system (the African Trypanosome, T. brucei) to study long-term, immune-evasive processes in a collaborative, strong immunology program at the German Cancer Research Center. Our initial results have been paradigm shifting.
T. brucei causes sleeping sickness in humans and related diseases in animals. Central to its immune-evasion strategy is the Variant Surface Glycoprotein (VSG) that forms a unique and extremely dense coat of ~10 million molecules carpeting the surface of the organism. The VSG coat elicits a robust antibody response that the parasite evades by accessing a large genetic repertoire of divergent VSGs and “switching” to a new (antigenically distinct) variant, resulting in long-term infection with observable peaks and valleys of parasitemia. These oscillations are the outcome of repeated cycles of antibody generation, parasite killing, and VSG switching: a process known as antigenic variation.
Our program is to mechanistically characterize the highly diverse VSGs and their complexes with antibodies. Initial successes of this “bottom-up” approach using X-ray crystallography have dramatically altered our view of T. brucei immune evasion with discoveries such as (1) major and unexpected structural divergence in the VSGs, (2) the identification of unanticipated and immune-modulatory post-translational modifications to the VSGs, vastly expanding their “epitope space”, (3) antibody-VSG co-crystal structures that have for the first time mapped the epitope surface accessibility of the coat, (4) VSG function beyond “mere” antigenic variation (e.g., binding biologically relevant substrates), (5) demonstration that the individual VSGs elicit unusual antibody repertoires that are highly focused on immunodominant epitopes, and (6) the translational development of a novel and powerful vaccine platform based on this surface coat and critically informed by protein structural considerations.
Technicians, students, and postdocs work within this background on projects to characterize VSGs (mapping their immunological “diversity space”) and antibody co-crystal structures with antigen. Techniques used in bench work include tissue culture, growth of eukaryotic microorganisms, large-scale protein purification and crystallization (both endogenous in the African trypanosome and recombinant, through bacterial expression), structural determination by X-ray crystallography and cryo-electron microscopy.
Photos of the lab
-

Image of multiple VSG structures -

DKFZ building on the campus of Heidelberg University -

Lab and friends out at the Christmas market in Heidelberg. -

X-ray diffractometer at DKFZ -

X-ray diffractometer at DKFZ -

Liquid N2 cryo-cooling for X-ray diffractometer at DKFZ -

X-ray diffraction image -

Laboratory space -
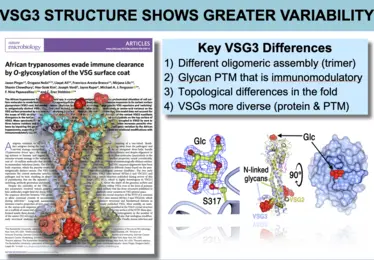
Images from publication -
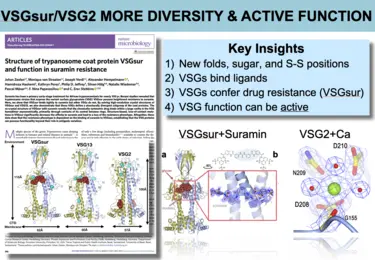
Images from the publication -
People and results from publication -

DKFZ main building on Heidelberg University campus -

Lab BBQ outing at the Grillhütte in Heidelberg -
Images from publication -

Visit from Director Baumann -
Images from publication -
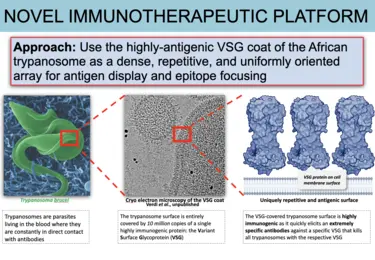
VAST immunotherapeutics platform -
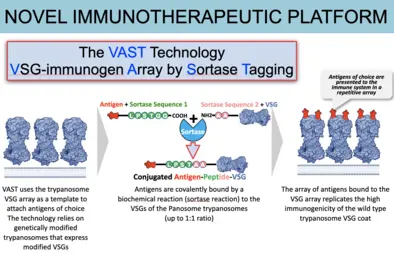
VAST immunotherapeutics platform -

Lab outing at Ketterwald (tree climbing and obstacle course) -

4.5 million EURO federal grant to DKFZ and Panosome spin-off
Team
-

Dr. Erec Stebbins
Head of Department
-

Dr. Tanja Isabell Kuhm
Postdoc
-

Caterina Canetta
Doctoral Student
-

Johan Philip Zeelen
Technical Engineer
-

Monique van Straaten
Technical Engineer
- Show profile

Christoph Tim Pufall
Administrative Assistant
Selected Publications
Dakovic S, Zeelen JP, Gkeka A, Chandra M, van Straaten M, Foti K, Zhong J, Vlachou EP, Aresta-Branco F, Verdi JP, Papavasiliou FN, Stebbins CE
Gkeka A, Aresta-Branco F, Triller G, Vlachou EP, van Straaten M, Lilic M, Olinares PDB, Perez K, Chait BT, Blatnik R, Ruppert T, Verdi JP, Stebbins CE, Papavasiliou FN
Chandra M, Dakovic S, Foti K, Zeelen JP, van Straaten M, Aresta-Branco F, Tihon E, Lübbehusen N, Ruppert T, Glover L, Papavasiliou FN, Stebbins CE
Triller G, Vlachou EP, Hashemi H, van Straaten M, Zeelen JP, Kelemen Y, Baehr C, Marker CL, Ruf S, Svirina A, Chandra M, Urban K, Gkeka A, Kruse S, Baumann A, Miller AK, Bartel M, Pravetoni M, Stebbins CE, Papavasiliou FN, Verdi JP
Get in touch with us
