Translational Medical Oncology
- Functional and Structural Genomics
- NCT

Prof. Dr. med. Stefan Fröhling
The aim of translational oncology is to transfer the results of cancer research into clinical application. This means that new findings on the development and progression of cancer should quickly benefit affected patients in the form of improved diagnosis and treatment options.

Our Research
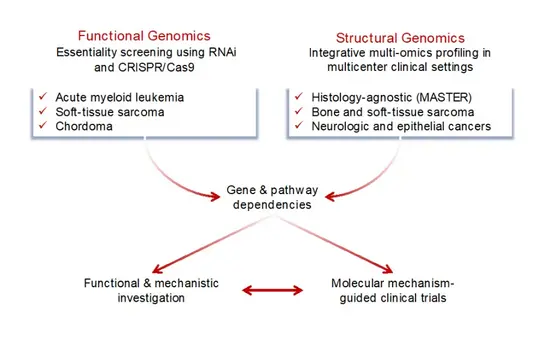
The Division of Translational Medical Oncology, established in 2019, has two overarching goals. First, investigators in the laboratory use functional and structural genetic approaches to gain insights into cancer biology that can be translated into novel diagnostic and therapeutic strategies. Second, clinically oriented investigators are working within prospective programs to generate comprehensive molecular portraits of large numbers of human cancers and use the results to inform clinical action – both at the level of individual patients and in the context of academically initiated trials – while feeding results back into exploratory research projects aimed at improving functional and mechanistic understanding of cancers. In keeping with these forward- and reverse-translational efforts at the interface between experimental cancer research and clinical application, which have a particular focus on the large and underserved group of rare cancers, the division's researchers represent multiple career paths and stages (postdoctoral scientists, Ph.D., M.Sc., and B.Sc. students; oncologists and M.D. students) and are supported by research technicians, study nurses, documentation staff, and dedicated administrative, data management, and scientific coordination teams.
Since its establishment, the division has been focusing on various aspects of biology-guided precision oncology and their clinical implementation and on the molecular pathogenesis and therapeutic targeting of rare cancers. More recently, additional strategic focus has been placed, and structurally supported, on the emerging areas of translational cancer epigenomics and tumor heterogeneity and treatment resistance.
Biology-Guided Precision Oncology
Efforts in biology-guided precision oncology can be divided into three focus areas represented by dedicated teams that have taken shape through the division's work over the last years. These relate to (a) the multidimensional characterization of human cancers within prospective observational trials (led by Peter Horak and Veronica Teleanu), (b) strategies for the scientific and clinical use of the resulting information layers and for making them available to the scientific community (led by Simon Kreutzfeldt), and (c) the translation of insights gained from clinically annotated multiomics datasets into molecularly stratified interventional trials (led by Christoph Heilig).
Multidimensional Tumor Characterization
Through collaboration with the Divisions of Molecular Genetics and Applied Bioinformatics, among others, and the Genomics and Proteomics and Omics IT and Data Management Core Facilities at DKFZ, the Molecular Precision Oncology Program of the National Center for Tumor Diseases (NCT) Heidelberg, the Division of Translational Medical Oncology at NCT Dresden, and all partner sites of the German Cancer Consortium (DKTK), the division has established a standardized and quality-controlled precision oncology workflow comprising (a) patient enrollment, (b) tissue processing and molecular profiling under accredited conditions, (c) bioinformatic analysis and biological curation of data, and (d) clinical decision-making in molecular tumor boards (MTBs) held several times a week. This pipeline has enabled the analysis of several thousand cancer genomes/exomes (including matched normal controls) and transcriptomes in prospective observational studies and the clinical use of the results.
The expertise in multiomics-based precision oncology is exemplified by the DKFZ/NCT/DKTK MASTER (Molecularly Aided Stratification for Tumor Eradication Research) program, focused on young adults with advanced malignancies of all entities and adults with rare cancers of all ages. This network was founded at DKFZ and NCT Heidelberg in 2013, expanded to NCT Dresden in 2015, and since 2016 also includes all DKTK sites (Berlin, Dresden, Essen/Düsseldorf, Frankfurt/Mainz, Freiburg, Munich, Tübingen) with their respective catchment areas. In total, well over 100 partners representing the entire spectrum of cancer patient care – from university hospitals to oncologists in private practice – have contributed to the program's success.
The MASTER program is an innovation driver for precision oncology in Germany regarding technology and structure. On the one hand, it has realized the clinical use of genome/exome and transcriptome sequencing from the outset (Cuppen E et al, 2022, JCO Precis Oncol 6:e2200245). This standard is constantly extended by new information layers (e.g., methylome, proteome, and single-cell profiles), and close collaborations exist with the National Network Genomic Medicine Lung Cancer and the Centers for Personalized Medicine in the state of Baden-Württemberg to translate these concepts into primary care structures. In addition, key structural elements of personalized oncology in Germany have been piloted and continuously developed, e.g., cross-site MTBs, standards for evidence grading, prioritization, and reporting of genetic variants in the clinical context, and new endpoints for precision oncology trials (Rieke D et al, 2022, BMC Med 20:367; Horak P et al, 2022, Genet Med 24:986-998; Horak P et al, 2022, Genes Chromosomes Cancer 61:303-313; Leichsenring J et al, 2019, Int J Cancer 145:2996-3010; Mock A et al, 2019, ESMO Open 4:e000583; Lier A et al, 2018, JCO Precis Oncol 2:1-13). Furthermore, the division is pursuing the establishment of an overarching quality management system with the aim of accrediting the entire workflow as a prerequisite for the use of whole-genome sequencing – and, prospectively, other comprehensive profiling approaches – in clinical cancer care.
Significant accomplishments of the MASTER program include, e.g., (a) the demonstration of the utility of comprehensive molecular profiling, including systematic cancer predisposition testing, for guiding therapeutic decisions in large cohorts of patients with rare malignancies (Horak P et al, 2021, Cancer Discov 11:2780-2795; Jahn A et al, 2022, Ann Oncol 33:1186-1199); (b) the finding of substantial diagnostic and therapeutic value of multiomics profiling in patients with carcinoma of unknown primary site (Möhrmann L et al, 2022, Nat Commun 13:4485); (c) the discovery of targetable gene fusions in patients with KRAS-wildtype pancreatic ductal adenocarcinoma (Heining C et al, 2018, Cancer Discov 8:1087-1095); and (d) the basis for mapping the landscape of genomic alterations such as chromothripsis across a broad spectrum of adult cancers (Voronina N et al, 2020, Nat Commun 11:2320, led by Aurélie Ernst, DKFZ Junior Research Group "Genome Instability in Tumors").
Finally, the division is intensely engaged in facilitating patient access to and participation in modern precision oncology through the MASTER program. This is reflected in the establishment of a patient advisory council, measures to explain the opportunities and risks of comprehensive molecular diagnostics to affected individuals and their families in non-expert language, and translational research projects in which patients are actively involved (Heilig CE et al, 2022, Eur J Cancer 172:107-118).
Data Science
The ongoing transformation of cancer care into data-driven precision oncology faces the problem of limited human and financial resources, leading to challenges in two areas. Clinical challenges relate to the scalability of precision oncology pipelines, the time required to evaluate clinical and associated molecular datasets, and the continuous improvement of data analysis quality despite increasing complexity. Scientific challenges are the accessibility of data and the speed of data analysis to expedite new discoveries. Based on the needs of the MASTER program, one of the leading precision oncology programs regarding the range of data collected and patients enrolled, the division pursues several projects to address these challenges. To provide precision oncology data resources to the community, it engages in (a) use case-driven modeling of patient data; (b) maintaining a precision oncology data thesaurus covering relevant taxonomic entities, e.g., drugs, therapies, and molecular alteration classes (Kreutzfeldt S et al, 2023, JCO Clin Cancer Inform 7:e2200147); (c) operating a database of molecular alteration-based evidence for treatment response; (d) developing algorithms and machine learning tools to predict treatment outcome from molecular alterations; and (e) developing the TEAPOT (Trials Extensively Annotated for Precision Oncology Therapies) clinical trial database.
As tools supporting precision oncology workflows, the division provides a dedicated Onkostar® environment for capturing clinical data in the context of precision oncology registries and develops the Knowledge Connector, an integrated, web-based platform for the semi-automated clinical assessment and presentation of multiomics data in MTBs. In addition to these strategically important own developments, the division's expertise has also been instrumental in the efforts of other comprehensive cancer centers in Germany and across Europe to build software solutions for automatically matching patients' molecular and clinical profiles with treatment recommendations, such as the Cancer Core Europe (CCE) Molecular Tumor Board Portal (Perera-Bel J et al, 2018, Genome Med 10:18; Borchert F et al, 2021, Brief Bioinform 22:1-17; Tamborero D et al, 2020, Nat Med 26:992-994; Tamborero D et al, 2022, Nat Cancer 3:251-261).
To enable mining of MASTER data for scientific purposes, the division maintains program-specific Tableau® and cBioportal for Cancer Genomics instances and provides an integrated database of all multiomics and clinical data from patients enrolled in MASTER for in-depth analyses using tools such as R and Python. Furthermore, to support the mission of GHGA (German Human Genome-Phenome Archive) of creating a secure national infrastructure for the use of human genome data, thus bridging the gap between research and medical care, the division made an early commitment to sharing the data generated in MASTER with the scientific community through GHGA. To improve treatment response prediction and develop methods for the design, conduct, and analysis of clinical trials, the division is involved in clinical initiatives spearheaded by NCT (Decision Support by Clustering based on Similarity Measures in Precision Oncology of Neuroendocrine Neoplasia and Sarcoma [DECISIONS], supported by the NCT Proof-of-Concept program) and CCE (Building Data-Rich Clinical Trials [DART], supported by the European Union).
Clinical Trials
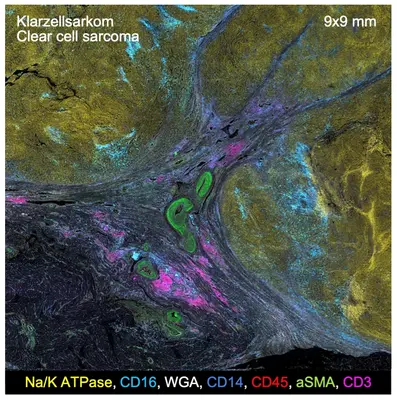
The clinical branch of the division aims to translate the results of multidimensional tumor profiling and laboratory-based cancer research into novel diagnostic and therapeutic approaches and, conversely, improve patient access to molecular diagnostics and innovative clinical trials. Together with the NCT Trial Center, the division develops and operates the NCT Precision Medicine in Oncology program, which currently includes four investigator-initiated, multicenter precision oncology studies that are open for enrollment (ClinicalTrials.gov Identifiers NCT03110744, NCT03127215, NCT04551521, NCT04410653 and Heilig CE et al, 2021, ESMO Open 6(6):100310). All trials are closely linked to MASTER, and two have yielded associated DKTK Joint Funding projects to investigate predictors of response or resistance to various cancer therapies, including conventional, molecularly targeted, and immunologic approaches. In addition, the division has a strong interest in applied sarcoma research focused on the multilayered characterization of rare subentities, the development of molecular predictors of treatment response, and the discovery of novel therapeutic targets. To this end, the division runs an outpatient clinic for patients with rare sarcomas to optimize diagnostic and therapeutic approaches and support research on these often poorly understood entities. Furthermore, it is significantly involved in SarcBOP (Sarcoma Biology and Outcomes Project, NCT04758325), a prospective registry study and biobank for sarcomas, and maintains interdisciplinary collaborations with the Departments of Radiation Oncology, Visceral Surgery, and Orthopedic Surgery at Heidelberg University Hospital, as well as with the Divisions of Applied Functional Genomics, Translational Pediatric Sarcoma Research, and Pediatric Neurooncology and the Junior Research Groups Soft-Tissue Sarcoma and Precision Sarcoma Research at DKFZ, among others. Finally, the division has played a leading role in evaluating the feasibility of comprehensive molecular profiling for clinical decision-making in adolescents and young adults with bone and soft-tissue sarcomas in an international setting within a prospective registry study by the EORTC (European Organisation for. Research and Treatment of Cancer; de Rojas T et al, 2020 Int J Cancer 147:1180-1184; Morfouace M et al, 2023, Eur J Cancer 178:216-226).
Molecular Pathogenesis and Therapeutic Targeting of Rare Cancers
The division has a strong interest in rare cancers, which collectively account for nearly 25% of all malignancies, as they are poorly understood and consequently have few treatment options available, resulting in a poorer prognosis than that of more common entities (van der Graaf WTA et al, 2022, Semin Cancer Biol 84:228-241). To address this challenge, the division uses structural and functional genomic approaches, with the ultimate goal of identifying targets for precision cancer therapy. On the one hand, we perform comprehensive genomic, transcriptomic, and epigenomic profiling of primary patient samples, e.g., in the MASTER program, which enrolls approximately 75% of patients with rare cancers, to capture the full spectrum of pathogenetically relevant and clinically actionable molecular alterations. On the other hand, we use large-scale RNA interference and, more recently, CRISPR screens to identify genotype-selective vulnerabilities in various cancer models. Candidate genes or pathways that emerge from these efforts are investigated functionally and mechanistically using appropriate model systems. Promising targets are taken forward to molecular mechanism-aware clinical trials.
Activities in the field of bone and soft-tissue sarcoma include (a) the investigation of the actionable genomic and transcriptomic landscapes of leiomyosarcoma and chordoma (Chudasama P et al, 2018, Nat Commun 9:144; Gröschel S et al, 2019, Nat Commun 10:1635), efforts that have led to a clinical trial; (b) the analysis of deregulated Hippo signaling in myxoid liposarcoma (Trautmann M et al, 2019, EMBO Mol Med 11:e9889; Berthold R et al, 2022, Oncogenesis 11:20); (c) the study of essential signaling pathways in soft-tissue sarcomas of the upper gastrointestinal tract (Mühlenberg T et al, 2019, Mol Cancer Ther 18:1985-1996; Heilig CE et al, 2020, Genes Chromosomes Cancer 59:601-608); and (d) application-oriented efforts such as a feasibility study of comprehensive molecular profiling in an international setting as part of an EORTC study, the development of a molecular predictor of response to pazopanib (Heilig CE et al, 2022, Eur J Cancer 172:107-118), the exploration of liquid biopsy techniques for the non-invasive detection of fusion-driven sarcomas (McConnell L et al, 2020, Cancers 12:3627), and contributions to sarcoma classification using DNA methylation profiling (Koelsche C et al, 2021, Nat Commun 12:498; Simon M et al, 2021, J Transl Med 19:204).
In the area of hematologic neoplasms, the division has (a) led efforts to identify RET-mediated autophagy suppression and aberrant expression of LIM kinases as targetable dependencies in acute myeloid leukemia (Rudat S et al, 2018, Leukemia 32:2189-2202; Jensen P et al, 2020, Leukemia 34:3173-3185) and (b) contributed to studies investigating myeloid transformation mediated by the transcription factor Cdx2 in vivo (Vu T et al, 2020, Nat Commun 11:3021) and characterizing the genomic landscape of T-cell acute lymphoblastic leukemia induced by gene therapy (Horak P et al, 2020, Leukemia 34:2785-2789). Achievements in the field of neurologic malignancies include the discovery of (a) activating ERBB2 mutations as a diagnostic parameter and therapeutic target in peripheral nervous system tumors (Ronellenfitsch M et al, 2020, J Clin Invest 130:2488-2495) and (b) a BRAF fusion oncoprotein with retained autoinhibitory domains in glioblastoma (Weinberg F et al, 2020, Oncogene 39:814-832). Finally, the division has contributed to several studies aimed at elucidating the molecular profiles and essential signaling pathways of rare epithelial cancers such as cholangiocellular carcinoma, pancreatoblastoma, and parathyroid carcinoma (Hoffmeister-Wittmann P et al, 2022, Liver Int 42:2855-2870; Sobol B et al, 2022, Int J Mol Sci 23:7850; Scherr AL et al, 2020, Cell Death Dis 11:875; Berger AK et al, 2020, Pancreatology 20:425-432; Reissig TM et al, 2022, Virchows Arch 481:265-272; Teleanu MV et al, 2023, Mol Oncol [Epub ahead of print]).
Translational Cancer Epigenomics
Molecular Classification and Risk-Adapted Therapy of Hematologic Malignancies
The division's activities in translational cancer epigenomics, led by Daniel Lipka, combine basic and translational research to develop novel approaches to cancer diagnosis, prognostication, and therapy. A particular focus is juvenile myelomonocytic leukemia (JMML), a rare myeloproliferative disease of early childhood characterized by constitutive RAS signaling and with a highly heterogeneous clinical course that is poorly captured by established clinical risk factors. The division has (a) spearheaded an international analysis of DNA methylation profiling as a tool for molecular categorization of JMML, (b) defined consensus molecular subclasses, and (c) developed and validated a machine learning algorithm that allows classification of prospective samples in a clinical setting (Schönung M et al, 2021, Clin Cancer Res 27:158-168) and has been applied, as part of a collaboration between DKFZ and the DKTK partner site Freiburg, to all newly diagnosed JMML patients enrolled in the EWOG-MDS (European Working Group on Myelodysplastic Syndromes in Childhood) registry since September 2021. In addition, this classifier has been adopted by the National Institutes of Health to make it available for patients in North America.
Based on case reports showing that JMML is sensitive to hypomethylating agents (HMAs), investigators in the division have contributed to a preclinical study showing that HMAs were effective in a JMML patient-derived xenograft model and targeted JMML stem cells (Krombholz CF et al, 2019, Leukemia 33:1805-1810). A subsequent clinical trial demonstrated that azacytidine was safe and effective in children with JMML, leading to its approval for this indication (Niemeyer CM et al, 2021, Blood Adv 5:2901-2908). A related approach demonstrated that DNMT3A-mutant acute myeloid leukemia preferentially responded to HMA treatment and provided insight into the underlying molecular mechanisms (Scheller M et al, 2021, Nat Cancer 2:527-544; Meier R et al, 2022, Blood Cancer J 12:122). Recent work led by the division suggests that HMAs should be explored as a new therapeutic option for patients with advanced systemic lupus erythematosus (Czeh M et al, 2022, J Immunol 208:358-370).
Using chronic lymphocytic leukemia as an example, the division's translational cancer epigenomics team has established a computational framework to distinguish differentiation-associated from disease-specific epigenetic patterns (Wierzbinska JA et al, 2020, Genome Med 12:29). Building on this work, the team has now performed a single-cell multiomics study to examine the hematopoietic stem and progenitor cell compartment of JMML patients. These efforts have led to the identification of oncofetal reprogramming as a central mechanism driving the aggressiveness of high-risk disease and the discovery of cell surface markers that are abnormally expressed on JMML stem cells and could serve as diagnostic markers and novel therapeutic targets.
Method Development and Integration Into Precision Oncology Workflows
The division has implemented array-based DNA methylation analysis using the Illumina Infinium EPIC Methylation BeadChip platform for all patients enrolled in the MASTER program since 2019. This information is currently being used to confirm clinical diagnoses and determine the likely origin of cancers of unknown primary site using a custom-trained pan-cancer tumor classifier and the published brain tumor and sarcoma methylation classifiers (Sill M et al, 2020, Hum Mol Genet 29:R205-R213). Furthermore, the division is co-developing novel DNA methylation biomarkers for treatment response prediction that can be used to inform recommendations by MTBs (Goeppert B et al, 2019, Hepatology 69:2091-2106; Niger M et al, 2022, Mol Oncol 16:2733-2746; Manoochehri M et al, 2023, Int J Cancer 152:1025-1035; Hey J et al, 2023, Int J Cancer 152:1226-1242).
In addition to expanding the application of established technologies, the division has a strong interest in developing new methods. These include a bisulfite-free approach for the simultaneous study of DNA methylation and mutation patterns at single-cell resolution (Niemöller C et al, 2021, Commun Biol 4:153) and protocols for minimally invasive detection of genetic and epigenetic tumor heterogeneity. The focus on new experimental methods is complemented by activities in the area of data analysis, e.g., the development of an online tool to support next-generation sequencing-based targeted DNA methylation analysis (Schönung M et al, 2021, Epigenetics 16:933-939; Mayakonda A et al, 2020, Bioinformatics 36:5524-5525).
Tumor Heterogeneity and Treatment Resistance
A research direction the division has recently taken concerns the highly dynamic area of intratumoral heterogeneity (ITH) and evolution caused by the stepwise acquisition of genetic and non-genetic alterations that promote relapse and treatment resistance. Specifically, investigators in the division are using bone and soft-tissue sarcomas with chimeric fusion genes as paradigmatic diseases to study ITH in cancers with defined clonal genetic drivers and translate the results into new treatment strategies. Clinically, the few effective therapies, lack of improvement in outcomes over decades, and predominantly young age at diagnosis underscore the need to decrease the years of life lost in sarcoma patients. From a biological perspective, fusion-driven sarcomas (FDS) appear particularly suitable for exploring ITH as fusion genes are entity-defining, truncal genetic features that can be inferred from routine diagnostics and exist throughout tumors' lifetimes, facilitating the investigation of divergent subclones and plasticity. Furthermore, FDS exhibit low genetic complexity, which will aid in identifying ITH-promoting factors and responses to the evolutionary pressure of therapy.

To investigate ITH in as many dimensions as possible, the division has taken a leading role in establishing the HEROES-AYA (Heterogeneity, Evolution, and Resistance in Oncogenic Fusion Gene-Expressing Sarcomas Affecting Adolescents and Young Adults) consortium (spokesperson, Stefan Fröhling; co-principal investigators, Stefan Pfister [Division of Pediatric Neurooncology] and Hanno Glimm [NCT Dresden]; partner sites, Heidelberg, Dresden, Berlin, Essen, Munich, Stuttgart, Tübingen). This multidisciplinary initiative is funded within the National Decade against Cancer with 19 million euros for five years from August 2022. It builds on INFORM (Individualized Therapy for Relapsed Malignancies in Childhood) and MASTER, DKFZ's internationally recognized precision oncology networks for children and young adults, and brings together experts in pediatric and adult sarcoma medicine, multiomic profiling, imaging, data science, preclinical modeling, clinical trial design, and patient advocacy to study FDS at diagnosis and the time of therapy resistance. Tumors are subjected to multilayered analyses at single-cell and spatial resolution to capture ITH at multiple levels. This will be complemented by in vitro and in vivo validation of newly identified vulnerabilities and the development of innovative clinical trials across age groups to improve outcomes for patients. Investigators in the division, in addition to their coordinating role, are mainly involved in (a) the genomic, transcriptomic, epigenomic, and proteomic characterization of FDS samples at the single-cell level and over time; (b) spatially resolved analyses of entire FDS ecosystems using newly devised proteomic profiling methods; (c) the development of new liquid biopsy techniques for minimally invasive monitoring of FDS evolution under the selective pressure of therapy; (d) the refinement of existing clinical workflows on which the experimental work is based; and (e) the involvement of patient representatives as research partners.
Team
Selected Publications
A. Jahn, A. Rump, T.J. Widmann, C. Heining, P. Horak, B. Hutter, N. Paramasivam, S. Uhrig, L. Gieldon, S. Drukewitz, A. Kübler, M. Bermudez, K. Hackmann, J. Porrmann, J. Wagner, M. Arlt, M. Franke, J. Fischer, Z. Kowalzyk, D. William, V. Weth, S. Oster, M. Fröhlich, J. Hüllein, C. Valle González, S. Kreutzfeldt, A. Mock, C.E. Heilig, D.B. Lipka, L. Möhrmann, D. Hanf, M. Oleś, V. Teleanu, M. Allgäuer, L. Ruhnke, O. Kutz, A. Knurr, A. Laßmann, V. Endris, O. Neumann, R. Penzel, K. Beck, D. Richter, U. Winter, S. Wolf, K. Pfütze, C. Geörg, B. Meißburger, I. Buchhalter, M. Augustin, W.E. Aulitzky, P. Hohenberger, M. Kroiss, P. Schirmacher, R.F. Schlenk, U. Keilholz, F. Klauschen, G. Folprecht, S. Bauer, J.T. Siveke, C.H. Brandts, T. Kindler, M. Boerries, A.L. Illert, N. von Bubnoff, P.J. Jost, K.H. Metzeler, M. Bitzer, K. Schulze-Osthoff, C. von Kalle, B. Brors, A. Stenzinger, W. Weichert, D. Hübschmann, S. Fröhling, H. Glimm, E. Schröck, B. Klink
Lino Möhrmann, Maximilian Werner, Małgorzata Oleś, Andreas Mock, Sebastian Uhrig, Arne Jahn, Simon Kreutzfeldt, Martina Fröhlich, Barbara Hutter, Nagarajan Paramasivam, Daniela Richter, Katja Beck, Ulrike Winter, Katrin Pfütze, Christoph E. Heilig, Veronica Teleanu, Daniel B. Lipka, Marc Zapatka, Dorothea Hanf, Catrin List, Michael Allgäuer, Roland Penzel, Gina Rüter, Ivan Jelas, Rainer Hamacher, Johanna Falkenhorst, Sebastian Wagner, Christian H. Brandts, Melanie Boerries, Anna L. Illert, Klaus H. Metzeler, C. Benedikt Westphalen, Alexander Desuki, Thomas Kindler, Gunnar Folprecht, Wilko Weichert, Benedikt Brors, Albrecht Stenzinger, Evelin Schröck, Daniel Hübschmann, Peter Horak, Christoph Heining, Stefan Fröhling, Hanno Glimm
Peter Horak, Christoph Heining, Simon Kreutzfeldt, Barbara Hutter, Andreas Mock, Jennifer Hüllein, Martina Fröhlich, Sebastian Uhrig, Arne Jahn, Andreas Rump, Laura Gieldon, Lino Möhrmann, Dorothea Hanf, Veronica Teleanu, Christoph E. Heilig, Daniel B. Lipka, Michael Allgäuer, Leo Ruhnke, Andreas Laßmann, Volker Endris, Olaf Neumann, Roland Penzel, Katja Beck, Daniela Richter, Ulrike Winter, Stephan Wolf, Katrin Pfütze, Christina Geörg, Bettina Meißburger, Ivo Buchhalter, Marinela Augustin, Walter E. Aulitzky, Peter Hohenberger, Matthias Kroiss, Peter Schirmacher, Richard F. Schlenk, Ulrich Keilholz, Frederick Klauschen, Gunnar Folprecht, Sebastian Bauer, Jens Thomas Siveke, Christian H. Brandts, Thomas Kindler, Melanie Boerries, Anna L. Illert, Nikolas von Bubnoff, Philipp J. Jost, Karsten Spiekermann, Michael Bitzer, Klaus Schulze-Osthoff, Christof von Kalle, Barbara Klink, Benedikt Brors, Albrecht Stenzinger, Evelin Schröck, Daniel Hübschmann, Wilko Weichert, Hanno Glimm, Stefan Fröhling
Michael W. Ronellenfitsch, Patrick N. Harter, Martina Kirchner, Christoph Heining, Barbara Hutter, Laura Gieldon, Jens Schittenhelm, Martin U. Schuhmann, Marcos Tatagiba, Gerhard Marquardt, Marlies Wagner, Volker Endris, Christian H. Brandts, Victor-Felix Mautner, Evelin Schröck, Wilko Weichert, Benedikt Brors, Andreas von Deimling, Michel Mittelbronn, Joachim P. Steinbach, David E. Reuss, Hanno Glimm, Albrecht Stenzinger, Stefan Fröhling
Get in touch with us
