Single-cell Open Lab
Dr. Jan-Philipp Mallm
The Single-cell Open Lab provides an infrastructure for processing cells, preparing libraries for single-cell sequencing as well as spatial omics. New users will be trained in most of our workflows so they can perform all experimental steps autonomously in the scOpenLab. We offer regular seminars to introduce new users to our portfolio. Systems can be booked online and a training session / detailed project discussion can be arranged. We also supports setting up new methods in collaborations.

Instrumentation
Available readouts
Chromium system (www.10xgenomics.com)
- scRNA-seq (3' / 5' counting)
- scVDJ-seq (+scRNA-seq)
- scATAC-seq
- multiome kit - RNA+ATAC
- CITE-seq & cell hashing
- numerous sample pooling strategies
- scDNA+RNA-seq (HIPSDR-seq)
- scChIP-seq + RNA-seq
The preparation of a clean single-cell suspension is key for a successful experiment. We can provide protocols / guidelines for dissociation, nuclei extraction from frozen tissue and nuclei preparation for scATAC-seq. All cell / nuclei suspensions can be analyzed with an automated cell counting system at the scOpenLab prior to loading.
Illumina Single Cell 3' RNA (https://www.illumina.com/products/by-type/sequencing-kits/library-prep-kits/single-cell-rna-prep.html)
Bead-based but microfluidics free workflow for generating 3' single-cell RNA-seq libraries. Cells and barcoding beads are mixed via several vortexing steps and RNA is eventually barcoded in a gel.
Tapestri (www.missionbio.com)
- targeted scDNA-seq
- SDR-seq
- protein + DNA-seq
For targeted single-cell sequencing we can provide access to the Tapestri (scDNA-seq) system. Panels for targeted amplification can be designed or directly purchased from the companies - for further information please visit their websites.
Combinatorial barcoding
Cells or nuclei can also be used a reaction compartments to barcode RNA in a cell-unique fashion. Two workflows are established in the scOpenLab from parse biosciences and scale biosciences (now part of 10X Genomics).
https://www.parsebiosciences.com/
Automized on plates
- scRNA-seq (SMART-seq 2.5, muta-seq and others)
- scHiC
- scWGBS
- G&T-seq
- scNMT-seq
- scWGS / scWES (PTA)
Automated workflows are conducted with the Bravo, Mosquito, C.wash and Mantis systems, depending on the reaction volume. Customized protocols can be programmed and run on all machines and we are happy to work with you for setting up those protocols.
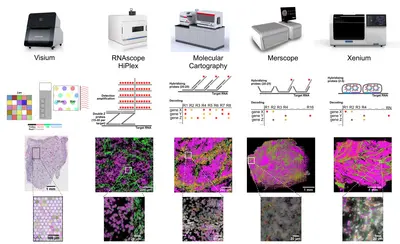
RNAscope HiPlex Assay
We offer test chemistry, guidance and experimental support for HiPlex Assays. Both fresh frozen und FFPE samples can be analysed with pre-defined targeted FISH probes and single-cell resolution. Staining and imaging equipment is available via our booking system.
10x Genomics Visium slide
While RNAscope provides single-cell resolution for up to 48 different pre-defined targets, visium sildes cover the whole transcriptome using spotted barcodes with a diameter 55 µm or with 2x2 µm square in case of visium HD. All steps including optimization, imaging, Cytassist transfer and library preparation can be done in our spatial lab.
Merfish
Merfish enables users to detect up to 500 transcripts in fresh frozen and FFPE samples. Subsequent antibody readouts can be performed for five protein targets using prelabeled secondary antibodies.
Xenium
The Xenium chemistry can detect up to 400 transcripts in fresh frozen and FFPE samples. Signal amplification via a rolling circle reaction allows the analysis of isoforms or fusions. Also 5k panels can be run on our system.
Each methodology has its merits (apart from the number of transcripts). If you are lost in space, we are happy to provide guidance in selecting the best technology for your project. Please discuss your spatial projects at an early stage with us - please do not order anything prior to a project discussion!
COMET by Lunaphore
We offer training and test antibodies to familiarize users with the seqIF workflow. Both fresh frozen und FFPE samples can be analysed. Staining and imaging is performed automatically by the system after successful panel optimization.
MACSima by Miltenyi
The MACSima allows detection of over 100 proteins via directly labelled antibodies. The fluorophore is either bleached or removed using the REAdye technology. Regular glass slides can be used in this workflow with diifferntly sized imaging chambers. We offer support in starting projects, testing antibodies and training to use this instrument.
Each methodology has its merits. If you are lost in space, we are happy to provide guidance in selecting the best technology for your project. Please discuss your spatial projects at an early stage with us - please do not order anything prior to a project discussion!
Available readouts
Single-cell sequencing
Chromium system (www.10xgenomics.com)
- scRNA-seq (3' / 5' counting)
- scVDJ-seq (+scRNA-seq)
- scATAC-seq
- multiome kit - RNA+ATAC
- CITE-seq & cell hashing
- numerous sample pooling strategies
- scDNA+RNA-seq (HIPSDR-seq)
- scChIP-seq + RNA-seq
The preparation of a clean single-cell suspension is key for a successful experiment. We can provide protocols / guidelines for dissociation, nuclei extraction from frozen tissue and nuclei preparation for scATAC-seq. All cell / nuclei suspensions can be analyzed with an automated cell counting system at the scOpenLab prior to loading.
Tapestri (www.missionbio.com)
- targeted scDNA-seq
- SDR-seq
- protein + DNA-seq
For targeted single-cell sequencing we can provide access to the Tapestri (scDNA-seq) system. Panels for targeted amplification can be designed or directly purchased from the companies - for further information please visit their websites.
Automized on plates
- scRNA-seq (SMART-seq 2.5, muta-seq and others)
- scHiC
- scWGBS
- G&T-seq
- scNMT-seq
- scWGS / scWES (PTA)
Automated workflows are conducted with the Bravo, Mosquito, C.wash and Mantis systems, depending on the reaction volume. Customized protocols can be programmed and run on all machines and we are happy to work with you for setting up those protocols.
Spatial transcriptomics
RNAscope HiPlex Assay
We offer test chemistry, guidance and experimental support for HiPlex Assays. Both fresh frozen und FFPE samples can be analysed with pre-defined targeted FISH probes and single-cell resolution. Staining and imaging equipment is available via our booking system.
10x Genomics Visium slide
While RNAscope provides single-cell resolution for up to 48 different pre-defined targets, visium sildes cover the whole transcriptome using spotted barcodes with a diameter 55 µm or with 2x2 µm square in case of visium HD. All steps including optimization, imaging, Cytassist transfer and library preparation can be done in our spatial lab.
Merfish
Merfish enables users to detect up to 960 transcripts in fresh frozen and FFPE samples. Subsequent antibody readouts can be performed for five protein targets using prelabeled secondary antibodies.
Xenium
The Xenium chemistry can detect up to 400 transcripts in fresh frozen and FFPE samples. Signal amplification via a rolling circle reaction allows the analysis of isoforms or fusions. Also 5k panels can be run on our system.
Spatial proteomics
COMET by Lunaphore
We offer training and test antibodies to familiarize users with the seqIF workflow. Both fresh frozen und FFPE samples can be analysed. Staining and imaging is performed automatically by the system after successful panel optimization.
MACSima by Miltenyi
The MACSima allows detection of over 100 proteins via directly labelled antibodies. The fluorophore is either bleached or removed using the REAdye technology.
Each methodology has its merits (apart from the number of transcripts / proteins). If you are lost in space, we are happy to provide guidance in selecting the best technology for your project. Please discuss your spatial projects at an early stage with us - please do not order anything prior to a project discussion!
How to get started
1) Visit our introductory seminar
For all new users we offer a single-cell sequencing seminar covering the different techniques, pros & cons, project design and lab logistics. The introduction is mandatory for all new scOpenLab users.
At the following dates the next mandatory introductions will take place:
- 11.12.2025 9:30 - 11:00 AM TP3 B4.101 - Single cell sequencing If you would like to participate, please send an e-mail to: scopenlab(at)dkfz.de (subject: introductory seminar)
- 05.02.2026 9:30 - 11:00 AM TP3 B4.101 - Single cell sequencing Please register here: https://indico.dkfz.de/event/1467/
- 12.02.2026 9:30 - 11:00 AM TP3 B4.101 - Spatial omics Please register here: https://indico.dkfz.de/event/1493/
- 21.05.2026 9:30 - 11:00 AM TP3 B4.101 - Single cell sequencing Please register here: https://indico.dkfz.de/event/1469/
- 17.09.2026 9:30 - 11:00 AM TP3 B4.101 - Single cell sequencing Please register here: https://indico.dkfz.de/event/1470/
2) Make an appointment with us to discuss your project in detail (optional but highly recommended)
3) Create an account for booking machines online: ppms link. You will also find a collection of protocols and further material on our ppms webpage.
4) Book machines for your first experiment and request assistance
For all new users we will offer an introduction on an individual basis for each machine / technique in the lab according to their project.
We offer some central chemistry for e.g. optimizations, SMART-seq, barcodes, spatial transcriptomics workflows - please refer to the order section in our booking system.
5) Run your experiment with a scOpenLab staff member
Team
-
Frau Katharina Bauer
-
Dr. Pooja Sant
-
Frau Laura Giese
-
Frau Denise Keitel
-
Dr. Michele Bortolomeazzi
-
Dr. Cindy Pamela Ulloa Guerrero
-
Frau Laura Schütze
-
Dr. Jan-Philipp Mallm
Selected Publications
Li X, Lebeaupin C, Kadianaki A, Druelle-Cedano C, Vesper N, Rennert C, Huguet-Pradell J, Gomez Ramos B, Fan C, Piecyk RS, Zizmare L, Ramadori P, Li L, Frick L, Qiu M, Zhang C, Martins Nascentes Melo L, Ranvir VP, Shen P, Hanselmann J, Kosla J, Fernández-Vaquero M, Vucur M, Baskaran P, Bao X, Coleman OI, Tang Y, Cetin M, Chen Z, Jang I, Del Prete S, Rahbari M, Zhang P, Pham TV, Hou Y, Sun A, Gu L, Kim LC, Rothermel U, Heide D, Ali A, Gallage S, Talvard-Balland N, Piqué-Gili M, Gris-Oliver A, Bevilacqua A, Schlicker L, Duffey A, Unger K, Szydlowska M, Hetzer J, Odom DT, Machauer T, Bucci D, Sant P, Lee JH, Rösler J, Meckelmann SW, Schreck J, Murray S, Simon MC, Nahnsen S, Schulze A, Ho PC, Jugold M, Breuhahn K, Mallm JP, Schirmacher P, Roth S, Rahbari N, Tschaharganeh DF, Roessler S, Goeppert B, Bengsch B, Andrieux G, Boerries M, Malek NP, Prinz M, Weber A, Zeiser R, Tamayo P, Bronsert P, Kurowski K, Thimme R, Yuan D, Carretero R, Luedde T, Pinyol R, Hartmann FJ, Karin M, Tasdogan A, Trautwein C, Mall M, Hofmann M, Llovet JM, Haller D, Kaufman RJ, Heikenwälder M.
Seufert I, Gerosa I, Varamogianni-Mamatsi V, Vladimirova A, Sen E, Mantz S, Rademacher A, Schumacher S, Liakopoulos P, Kolovos P, Anders S, Mallm JP, Papantonis A, Rippe K.
Hartmann M, Schönung M, Rajak J, Maurer V, Hai L, Bauer K, Hakobyan M, Stäble S, Langstein J, Jardine L, Roelz R, Bohler S, Khabirova E, Maag AH, Vonficht D, Lebrecht D, Bernt KM, Tan K, Chen C, Alikarami F, Meyer J, Wang J, Boch T, Flore V, Lutsik P, Milsom MD, Raffel S, Buske C, Haas S, Haniffa M, Mallm JP, Behjati S, Bonder MJ, Frohling S, Stieglitz E, Niemeyer CM, Hey J, Flotho C, Plass C, Erlacher M, Schlesner M, Lipka DB.
Functional and molecular analyses reveal impaired HSPCs in Multiple Myeloma patients post-induction.
Baumhardt TM, Amoah A, Hoenicka M, Liebold A, Sakk V, Soller K, Vollmer A, Kull M, Kronke J, Mallm JP, Geiger H, Mulaw M.
Staehle HF, Koellerer C, Staehle AM, Schulze J, Eble P, Müller A, Zell F, Müller JM, Perner F, Attia A, Mallm JP, Pozdnyakova O, Rippe K, Brors B, Feuerbach L, Imbusch CD, Metzger E, Schüle R, Pahl HL, Jutzi JS.
Llaó-Cid L, Wong J, Fernandez Botana I, Paul Y, Wierz M, Pilger LM, Floerchinger A, Tan CL, Gonder S, Pagano G, Chazotte M, Bestak K, Schifflers C, Iskar M, Roider T, Czernilofsky F, Bruch PM, Mallm JP, Cosma A, Campton DE, Gerhard-Hartmann E, Rosenwald A, Colomer D, Campo E, Schapiro D, Green EW, Dietrich S, Lichter P, Moussay E, Paggetti J, Zapatka M, Seiffert M.
Rademacher A, Huseynov A, Bortolomeazzi M, Wille SJ, Schumacher S, Sant P, Keitel D, Okonechnikov K, Ghasemi DR, Pajtler KW, Mallm JP, Rippe K.
Okonechnikov K, Joshi P, Körber V, Rademacher A, Bortolomeazzi M, Mallm JP, Vaillant J, da Silva PBG, Statz B, Sepp M, Sarropoulos I, Yamada T, Wittmann A, Schramm K, Blattner-Johnson M, Fiesel P, Jones B, Jäger N, Milde T, Pajtler KW, van Tilburg CM, Witt O, Bochennek K, Weber KJ, Nonnenmacher L, Reimann C, Ghasemi DR, Schüller U, Mynarek M, Rutkowski S, Jones DTW, Korshunov A, Rippe K, Westermann F, Thongjuea S, Höfer T, Kaessmann H, Kutscher LM, Pfister SM.
Ernst KJ, Okonechnikov K, Bageritz J, Perera AA, Mallm JP, Wittmann A, Maaß KK, Leible S, Boutros M, Pfister SM, Zuckermann M, Jones DTW.
Thiel V, Renders S, Panten J, Dross N, Bauer K, Azorin D, Henriques V, Vogel V, Klein C, Leppä AM, Barriuso Ortega I, Schwickert J, Ourailidis I, Mochayedi J, Mallm JP, Müller-Tidow C, Monyer H, Neoptolemos J, Hackert T, Stegle O, Odom DT, Offringa R, Stenzinger A, Winkler F, Sprick M, Trumpp A.
Lutz R, Poos AM, Solé-Boldo L, John L, Wagner J, Prokoph N, Baertsch MA, Vonficht D, Palit S, Brobeil A, Mechtersheimer G, Hildenbrand N, Hemmer S, Steiger S, Horn S, Pepke W, Spranz DM, Rehnitz C, Sant P, Mallm JP, Friedrich MJ, Reichert P, Huhn S, Trumpp A, Rippe K, Haghverdi L, Fröhling S, Müller-Tidow C, Hübschmann D, Goldschmidt H, Willimsky G, Sauer S, Raab MS, Haas S, Weinhold N.
Smirnov P, Przybilla MJ, Simovic-Lorenz M, Parra RG, Susak H, Ratnaparkhe M, Wong JK, Körber V, Mallm JP, Philippos G, Sill M, Kolb T, Kumar R, Casiraghi N, Okonechnikov K, Ghasemi DR, Maaß KK, Pajtler KW, Jauch A, Korshunov A, Höfer T, Zapatka M, Pfister SM, Huber W, Stegle O, Ernst A.
Gehrs S, Jakab M, Gutjahr E, Gu Z, Weichenhan D, Mallm JP, Mogler C, Schlesner M, Plass C, Schlereth K, Augustin HG.
Otoničar J, Lazareva O, Mallm JP, Simovic-Lorenz M, Philippos G, Sant P, Parekh U, Hammann L, Li A, Yildiz U, Marttinen M, Zaugg J, Noh KM, Stegle O, Ernst A.
Kats I, Simovic-Lorenz M, Schreiber HS, Sant P, Mallm JP, Körber V, Li A, Velmurugan P, Heuer S, Kües L, Devens F, Sill M, Jugold M, Moustafa M, Abdollahi A, Winkler F, Korshunov A, Pfister SM, Stegle O, Ernst A.
Smirnov P, Przybilla MJ, Simovic-Lorenz M, Parra RG, Susak H, Ratnaparkhe M, Wong JK, Körber V, Mallm JP, Philippos G, Sill M, Kolb T, Kumar R, Casiraghi N, Okonechnikov K, Ghasemi DR, Maaß KK, Pajtler KW, Jauch A, Korshunov A, Höfer T, Zapatka M, Pfister SM, Huber W, Stegle O, Ernst A.
Ghasemi DR, Okonechnikov K, Rademacher A, Tirier S, Maass KK, Schumacher H, Joshi P, Gold MP, Sundheimer J, Statz B, Rifaioglu AS, Bauer K, Schumacher S, Bortolomeazzi M, Giangaspero F, Ernst KJ, Clifford SC, Saez-Rodriguez J, Jones DTW, Kawauchi D, Fraenkel E, Mallm JP, Rippe K, Korshunov A, Pfister SM, Pajtler KW.
Man KH, Wu Y, Gao Z, Spreng AS, Keding J, Mangei J, Boskovic P, Mallm JP, Liu HK, Imbusch CD, Lichter P, Radlwimmer B.
Roessner PM, Seufert I, Chapaprieta V, Jayabalan R, Briesch H, Massoni-Badosa R, Boskovic P, Benckendorff J, Roider T, Arseni L, Coelho M, Chakraborty S, Vaca AM, Sivina M, Muckenhuber M, Rodriguez-Rodriguez S, Bonato A, Herbst SA, Zapatka M, Sun C, Kretzmer H, Naake T, Bruch PM, Czernilofsky F, Ten Hacken E, Schneider M, Helm D, Yosifov DY, Kauer J, Danilov AV, Bewarder M, Heyne K, Schneider C, Stilgenbauer S, Wiestner A, Mallm JP, Burger JA, Efremov DG, Lichter P, Dietrich S, Martin-Subero JI, Rippe K, Seiffert M.
Bundschuh C, Weidner N, Klein J, Rausch T, Azevedo N, Telzerow A, Mallm JP, Kim H, Steiger S, Seufert I, Börner K, Bauer K, Hübschmann D, Jost KL, Parthé S, Schnitzler P, Boutros M, Rippe K, Müller B, Bartenschlager R, Kräusslich HG, Benes V.
Wang C, Sun M, Shao C, Schlicker L, Zhuo Y, Harim Y, Peng T, Tian W, Stöffler N, Schneider M, Helm D, Chu Y, Fu B, Jin X, Mallm JP, Mall M, Wu Y, Schulze A, Liu HK.
Ghasemi DR, Okonechnikov K, Rademacher A, Tirier S, Maass KK, Schumacher H, Joshi P, Gold MP, Sundheimer J, Statz B, Rifaioglu AS, Bauer K, Schumacher S, Bortolomeazzi M, Giangaspero F, Ernst KJ, Clifford SC, Saez-Rodriguez J, Jones DTW, Kawauchi D, Fraenkel E, Mallm JP, Rippe K, Korshunov A, Pfister SM, Pajtler KW.
Schuster LC, Syed AP, Tirier SM, Steiger S, Seufert I, Becker H, Duque-Afonso J, Ma T, Ogawa S, Mallm JP, Lübbert M, Rippe K.
Piroeva KV, McDonald C, Xanthopoulos C, Fox C, Clarkson CT, Mallm JP, Vainshtein Y, Ruje L, Klett LC, Stilgenbauer S, Mertens D, Kostareli E, Rippe K, Teif VB.
Wong JKL, Jassowicz L, Herold-Mende C, Seiffert M, Mallm JP, Lichter P, Zapatka M.
John L, Poos AM, Brobeil A, Schinke C, Huhn S, Prokoph N, Lutz R, Wagner B, Zangari M, Tirier SM, Mallm JP, Schumacher S, Vonficht D, Solé-Boldo L, Quick S, Steiger S, Przybilla MJ, Bauer K, Baumann A, Hemmer S, Rehnitz C, Lückerath C, Sachpekidis C, Mechtersheimer G, Haberkorn U, Dimitrakopoulou-Strauss A, Reichert P, Barlogie B, Müller-Tidow C, Goldschmidt H, Hillengass J, Rasche L, Haas SF, van Rhee F, Rippe K, Raab MS, Sauer S, Weinhold N.
Poos AM, Prokoph N, Przybilla MJ, Mallm JP, Steiger S, Seufert I, John L, Tirier SM, Bauer K, Baumann A, Rohleder J, Munawar U, Rasche L, Kortüm KM, Giesen N, Reichert P, Huhn S, Müller-Tidow C, Goldschmidt H, Stegle O, Raab MS, Rippe K, Weinhold N.
Sant P, Rippe K, Mallm JP.
Muckenhuber M, Seufert I, Müller-Ott K, Mallm JP, Klett LC, Knotz C, Hechler J, Kepper N, Erdel F, Rippe K.
Montserrat-Vazquez S, Ali NJ, Matteini F, Lozano J, Zhaowei T, Mejia-Ramirez E, Marka G, Vollmer A, Soller K, Sacma M, Sakk V, Mularoni L, Mallm JP, Plass M, Zheng Y, Geiger H, Florian MC.
Solé-Boldo L, Raddatz G, Gutekunst J, Gilliam O, Bormann F, Liberio MS, Hasche D, Antonopoulos W, Mallm JP, Lonsdorf AS, Rodríguez-Paredes M, Lyko F.
Bogeska R, Mikecin AM, Kaschutnig P, Fawaz M, Büchler-Schäff M, Le D, Ganuza M, Vollmer A, Paffenholz SV, Asada N, Rodriguez-Correa E, Frauhammer F, Buettner F, Ball M, Knoch J, Stäble S, Walter D, Petri A, Carreño-Gonzalez MJ, Wagner V, Brors B, Haas S, Lipka DB, Essers MAG, Weru V, Holland-Letz T, Mallm JP, Rippe K, Krämer S, Schlesner M, McKinney Freeman S, Florian MC, King KY, Frenette PS, Rieger MA, Milsom MD.
Hageb A, Thalheim T, Nattamai KJ, Möhrle B, Saçma M, Sakk V, Thielecke L, Cornils K, Grandy C, Port F, Gottschalk KE, Mallm JP, Glauche I, Galle J, Mulaw MA, Geiger H.
Tirier SM, Mallm JP, Steiger S, Poos AM, Awwad MHS, Giesen N, Casiraghi N, Susak H, Bauer K, Baumann A, John L, Seckinger A, Hose D, Müller-Tidow C, Goldschmidt H, Stegle O, Hundemer M, Weinhold N, Raab MS, Rippe K.
Derrien J, Guérin-Charbonnel C, Gaborit V, Campion L, Devic M, Douillard E, Roi N, Avet-Loiseau H, Decaux O, Facon T, Mallm JP, Eils R, Munshi NC, Moreau P, Herrmann C, Magrangeas F, Minvielle S.
Dudek M, Pfister D, Donakonda S, Filpe P, Schneider A, Laschinger M, Hartmann D, Hüser N, Meiser P, Bayerl F, Inverso D, Wigger J, Sebode M, Öllinger R, Rad R, Hegenbarth S, Anton M, Guillot A, Bowman A, Heide D, Müller F, Ramadori P, Leone V, Garcia-Caceres C, Gruber T, Seifert G, Kabat AM, Mallm JP, Reider S, Effenberger M, Roth S, Billeter AT, Müller-Stich B, Pearce EJ, Koch-Nolte F, Käser R, Tilg H, Thimme R, Boettler T, Tacke F, Dufour JF, Haller D, Murray PJ, Heeren R, Zehn D, Böttcher JP, Heikenwälder M, Knolle PA.
Reich M, Spomer L, Klindt C, Fuchs K, Stindt J, Deutschmann K, Höhne J, Liaskou E, Hov JR, Karlsen TH, Beuers U, Verheij J, Ferreira-Gonzalez S, Hirschfield G, Forbes SJ, Schramm C, Esposito I, Nierhoff D, Fickert P, Fuchs CD, Trauner M, García-Beccaria M, Gabernet G, Nahnsen S, Mallm JP, Vogel M, Schoonjans K, Lautwein T, Köhrer K, Häussinger D, Luedde T, Heikenwalder M, Keitel V.
Dudek M, Pfister D, Donakonda S, Filpe P, Schneider A, Laschinger M, Hartmann D, Hüser N, Meiser P, Bayerl F, Inverso D, Wigger J, Sebode M, Öllinger R, Rad R, Hegenbarth S, Anton M, Guillot A, Bowman A, Heide D, Müller F, Ramadori P, Leone V, Garcia-Caceres C, Gruber T, Seifert G, Kabat AM, Mallm JP, Reider S, Effenberger M, Roth S, Billeter AT, Müller-Stich B, Pearce EJ, Koch-Nolte F, Käser R, Tilg H, Thimme R, Boettler T, Tacke F, Dufour JF, Haller D, Murray PJ, Heeren R, Zehn D, Böttcher JP, Heikenwälder M, Knolle PA.
Pfister D, Núñez NG, Pinyol R, Govaere O, Pinter M, Szydlowska M, Gupta R, Qiu M, Deczkowska A, Weiner A, Müller F, Sinha A, Friebel E, Engleitner T, Lenggenhager D, Moncsek A, Heide D, Stirm K, Kosla J, Kotsiliti E, Leone V, Dudek M, Yousuf S, Inverso D, Singh I, Teijeiro A, Castet F, Montironi C, Haber PK, Tiniakos D, Bedossa P, Cockell S, Younes R, Vacca M, Marra F, Schattenberg JM, Allison M, Bugianesi E, Ratziu V, Pressiani T, D'Alessio A, Personeni N, Rimassa L, Daly AK, Scheiner B, Pomej K, Kirstein MM, Vogel A, Peck-Radosavljevic M, Hucke F, Finkelmeier F, Waidmann O, Trojan J, Schulze K, Wege H, Koch S, Weinmann A, Bueter M, Rössler F, Siebenhüner A, De Dosso S, Mallm JP, Umansky V, Jugold M, Luedde T, Schietinger A, Schirmacher P, Emu B, Augustin HG, Billeter A, Müller-Stich B, Kikuchi H, Duda DG, Kütting F, Waldschmidt DT, Ebert MP, Rahbari N, Mei HE, Schulz AR, Ringelhan M, Malek N, Spahn S, Bitzer M, Ruiz de Galarreta M, Lujambio A, Dufour JF, Marron TU, Kaseb A, Kudo M, Huang YH, Djouder N, Wolter K, Zender L, Marche PN, Decaens T, Pinato DJ, Rad R, Mertens JC, Weber A, Unger K, Meissner F, Roth S, Jilkova ZM, Claassen M, Anstee QM, Amit I, Knolle P, Becher B,…
Roider T, Seufert J, Uvarovskii A, Frauhammer F, Bordas M, Abedpour N, Stolarczyk M, Mallm JP, Herbst SA, Bruch PM, Balke-Want H, Hundemer M, Rippe K, Goeppert B, Seiffert M, Brors B, Mechtersheimer G, Zenz T, Peifer M, Chapuy B, Schlesner M, Müller-Tidow C, Fröhling S, Huber W, Anders S, Dietrich S.
Vu T, Straube J, Porter AH, Bywater M, Song A, Ling V, Cooper L, Pali G, Bruedigam C, Jacquelin S, Green J, Magor G, Perkins A, Chalk AM, Walkley CR, Heidel FH, Mukhopadhyay P, Cloonan N, Gröschel S, Mallm JP, Fröhling S, Scholl C, Lane SW.
Solé-Boldo L, Raddatz G, Schütz S, Mallm JP, Rippe K, Lonsdorf AS, Rodríguez-Paredes M, Lyko F.
Wierzbinska JA, Toth R, Ishaque N, Rippe K, Mallm JP, Klett LC, Mertens D, Zenz T, Hielscher T, Seifert M, Küppers R, Assenov Y, Lutsik P, Stilgenbauer S, Roessner PM, Seiffert M, Byrd J, Oakes CC, Plass C, Lipka DB.
Delacher M, Imbusch CD, Hotz-Wagenblatt A, Mallm JP, Bauer K, Simon M, Riegel D, Rendeiro AF, Bittner S, Sanderink L, Pant A, Schmidleithner L, Braband KL, Echtenachter B, Fischer A, Giunchiglia V, Hoffmann P, Edinger M, Bock C, Rehli M, Brors B, Schmidl C, Feuerer M.
Al-Ali R, Bauer K, Park JW, Al Abdulla R, Fermi V, von Deimling A, Herold-Mende C, Mallm JP, Herrmann C, Wick W, Turcan Ş.
Saçma M, Pospiech J, Bogeska R, de Back W, Mallm JP, Sakk V, Soller K, Marka G, Vollmer A, Karns R, Cabezas-Wallscheid N, Trumpp A, Méndez-Ferrer S, Milsom MD, Mulaw MA, Geiger H, Florian MC.
Mallm JP, Windisch P, Biran A, Gal Z, Schumacher S, Glass R, Herold-Mende C, Meshorer E, Barbus M, Rippe K.
Tirier SM, Park J, Preußer F, Amrhein L, Gu Z, Steiger S, Mallm JP, Krieger T, Waschow M, Eismann B, Gut M, Gut IG, Rippe K, Schlesner M, Theis F, Fuchs C, Ball CR, Glimm H, Eils R, Conrad C.
Mallm JP, Iskar M, Ishaque N, Klett LC, Kugler SJ, Muino JM, Teif VB, Poos AM, Großmann S, Erdel F, Tavernari D, Koser SD, Schumacher S, Brors B, König R, Remondini D, Vingron M, Stilgenbauer S, Lichter P, Zapatka M, Mertens D, Rippe K.
Becker H, Greve G, Kataoka K, Mallm JP, Duque-Afonso J, Ma T, Niemöller C, Pantic M, Duyster J, Cleary ML, Schüler J, Rippe K, Ogawa S, Lübbert M.
Kalamakis G, Brüne D, Ravichandran S, Bolz J, Fan W, Ziebell F, Stiehl T, Catalá-Martinez F, Kupke J, Zhao S, Llorens-Bobadilla E, Bauer K, Limpert S, Berger B, Christen U, Schmezer P, Mallm JP, Berninger B, Anders S, Del Sol A, Marciniak-Czochra A, Martin-Villalba A.
Erkek S, Johann PD, Finetti MA, Drosos Y, Chou HC, Zapatka M, Sturm D, Jones DTW, Korshunov A, Rhyzova M, Wolf S, Mallm JP, Beck K, Witt O, Kulozik AE, Frühwald MC, Northcott PA, Korbel JO, Lichter P, Eils R, Gajjar A, Roberts CWM, Williamson D, Hasselblatt M, Chavez L, Pfister SM, Kool M.
Zirkel A, Nikolic M, Sofiadis K, Mallm JP, Brackley CA, Gothe H, Drechsel O, Becker C, Altmüller J, Josipovic N, Georgomanolis T, Brant L, Franzen J, Koker M, Gusmao EG, Costa IG, Ullrich RT, Wagner W, Roukos V, Nürnberg P, Marenduzzo D, Rippe K, Papantonis A.
Delacher M, Imbusch CD, Weichenhan D, Breiling A, Hotz-Wagenblatt A, Träger U, Hofer AC, Kägebein D, Wang Q, Frauhammer F, Mallm JP, Bauer K, Herrmann C, Lang PA, Brors B, Plass C, Feuerer M.
Brocks D, Schmidt CR, Daskalakis M, Jang HS, Shah NM, Li D, Li J, Zhang B, Hou Y, Laudato S, Lipka DB, Schott J, Bierhoff H, Assenov Y, Helf M, Ressnerova A, Islam MS, Lindroth AM, Haas S, Essers M, Imbusch CD, Brors B, Oehme I, Witt O, Lübbert M, Mallm JP, Rippe K, Will R, Weichenhan D, Stoecklin G, Gerhäuser C, Oakes CC, Wang T, Plass C.
Delacher M, Imbusch CD, Weichenhan D, Breiling A, Hotz-Wagenblatt A, Träger U, Hofer AC, Kägebein D, Wang Q, Frauhammer F, Mallm JP, Bauer K, Herrmann C, Lang PA, Brors B, Plass C, Feuerer M.
Brocks D, Schmidt CR, Daskalakis M, Jang HS, Shah NM, Li D, Li J, Zhang B, Hou Y, Laudato S, Lipka DB, Schott J, Bierhoff H, Assenov Y, Helf M, Ressnerova A, Islam MS, Lindroth AM, Haas S, Essers M, Imbusch CD, Brors B, Oehme I, Witt O, Lübbert M, Mallm JP, Rippe K, Will R, Weichenhan D, Stoecklin G, Gerhäuser C, Oakes CC, Wang T, Plass C.
Teif VB, Mallm JP, Sharma T, Mark Welch DB, Rippe K, Eils R, Langowski J, Olins AL, Olins DE.
Molitor J, Mallm JP, Rippe K, Erdel F.
Bauer T, Trump S, Ishaque N, Thürmann L, Gu L, Bauer M, Bieg M, Gu Z, Weichenhan D, Mallm JP, Röder S, Herberth G, Takada E, Mücke O, Winter M, Junge KM, Grützmann K, Rolle-Kampczyk U, Wang Q, Lawerenz C, Borte M, Polte T, Schlesner M, Schanne M, Wiemann S, Geörg C, Stunnenberg HG, Plass C, Rippe K, Mizuguchi J, Herrmann C, Eils R, Lehmann I.
Mattout A, Aaronson Y, Sailaja BS, Raghu Ram EV, Harikumar A, Mallm JP, Sim KH, Nissim-Rafinia M, Supper E, Singh PB, Sze SK, Gasser SM, Rippe K, Meshorer E.
Junge KM, Bauer T, Geissler S, Hirche F, Thürmann L, Bauer M, Trump S, Bieg M, Weichenhan D, Gu L, Mallm JP, Ishaque N, Mücke O, Röder S, Herberth G, Diez U, Borte M, Rippe K, Plass C, Hermann C, Stangl GI, Eils R, Lehmann I.
Mallm JP, Rippe K.
Campos B, Weisang S, Osswald F, Ali R, Sedlmeier G, Bageritz J, Mallm JP, Hartmann C, von Deimling A, Popanda O, Goidts V, Plass C, Unterberg A, Schmezer P, Burhenne J, Herold-Mende C.
Yearim A, Gelfman S, Shayevitch R, Melcer S, Glaich O, Mallm JP, Nissim-Rafinia M, Cohen AH, Rippe K, Meshorer E, Ast G.
Hick M, Herrmann U, Weyer SW, Mallm JP, Tschäpe JA, Borgers M, Mercken M, Roth FC, Draguhn A, Slomianka L, Wolfer DP, Korte M, Müller UC.
Hick M, Herrmann U, Weyer SW, Mallm JP, Tschäpe JA, Borgers M, Mercken M, Roth FC, Draguhn A, Slomianka L, Wolfer DP, Korte M, Müller UC.
Müller-Ott K, Erdel F, Matveeva A, Mallm JP, Rademacher A, Hahn M, Bauer C, Zhang Q, Kaltofen S, Schotta G, Höfer T, Rippe K.
Teif VB, Beshnova DA, Vainshtein Y, Marth C, Mallm JP, Höfer T, Rippe K.
Jäger N, Schlesner M, Jones DT, Raffel S, Mallm JP, Junge KM, Weichenhan D, Bauer T, Ishaque N, Kool M, Northcott PA, Korshunov A, Drews RM, Koster J, Versteeg R, Richter J, Hummel M, Mack SC, Taylor MD, Witt H, Swartman B, Schulte-Bockholt D, Sultan M, Yaspo ML, Lehrach H, Hutter B, Brors B, Wolf S, Plass C, Siebert R, Trumpp A, Rippe K, Lehmann I, Lichter P, Pfister SM, Eils R.
Suv4-20h2 mediates chromatin compaction and is important for cohesin recruitment to heterochromatin.
Hahn M, Dambacher S, Dulev S, Kuznetsova AY, Eck S, Wörz S, Sadic D, Schulte M, Mallm JP, Maiser A, Debs P, von Melchner H, Leonhardt H, Schermelleh L, Rohr K, Rippe K, Storchova Z, Schotta G.
Teif VB, Erdel F, Beshnova DA, Vainshtein Y, Mallm JP, Rippe K.
Teif VB, Vainshtein Y, Caudron-Herger M, Mallm JP, Marth C, Höfer T, Rippe K.
Caudron-Herger M, Müller-Ott K, Mallm JP, Marth C, Schmidt U, Fejes-Tóth K, Rippe K.
Mallm JP, Tschäpe JA, Hick M, Filippov MA, Müller UC.
Plaimas K, Mallm JP, Oswald M, Svara F, Sourjik V, Eils R, König R.
Fatumo S, Plaimas K, Mallm JP, Schramm G, Adebiyi E, Oswald M, Eils R, König R.
Get in touch with us