Brain Tumor Translational Targets
- Functional and Structural Genomics

Dr. Violaine Goidts
Due to its location, aggressiveness and diffuse growth pattern, glioblastoma therapy is tremendously challenging and advances in the field are urgently needed.

Our Research
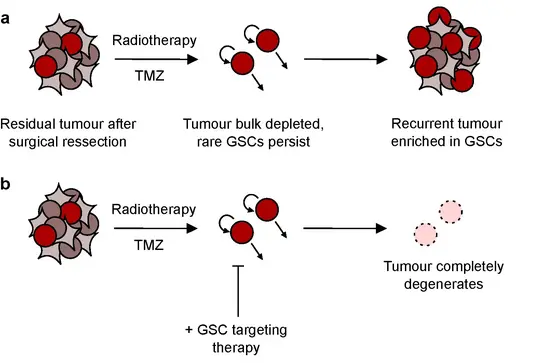
Glioblastoma stem-like cells (GSCs), which play a crucial role in tumor relapse, exhibit strong resistance to conventional chemo- and radiotherapy due to their robust self-renewal capacity. The primary goal of the research group is to identify molecular dependencies in GSCs to uncover novel therapeutic targets and improve patient outcomes.
Our main strategy involves functional screens using gene knock-down and fungal extract libraries to identify new candidate molecules to treat for glioblastoma patients. We integrate in silico data from high-throughput -OMICS technologies to our functional screen results to shed light on how to achieve better patient stratification and we design combinatorial treatments to optimize current treatment efficacy. More recently, our team has also developed a single-cell strategy to assess drug penetration in the brain and has begun exploring nanoparticle-based drug delivery systems to improve therapeutic efficiency.
Projects
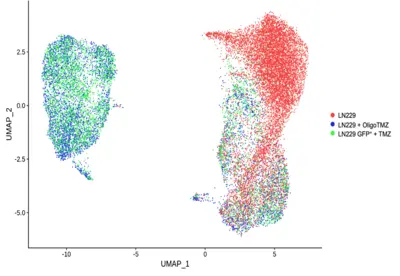
Overcoming drug resistance remains a major challenge in treating aggressive cancers. While advances in single-cell sequencing and spatial transcriptomics have improved our understanding of cancer therapies, current technologies cannot determine whether a drug has successfully penetrated specific cells or tissues or assess its precise effects. To address this, we are developing an innovative DNA barcoding technology for small molecules, enabling the detection and quantification of drugs within single-cell RNA sequencing and spatial transcriptomics data. This approach aims to revolutionize how we study drug efficacy and resistance at the single-cell level.
PFKFB4 (6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase 4) is a key regulator of glycolysis and is selectively overexpressed in various cancers, including glioblastoma, prostate, breast, and lung cancers, while being nearly absent in normal tissues. Our research has demonstrated that PFKFB4 plays a critical role in tumor metabolism, survival under hypoxic conditions, and drug resistance by supporting glioma stem-like cell survival and interacting with proteins like FBXO28 to regulate HIF-1α degradation. Silencing PFKFB4 significantly impairs tumor growth in preclinical models, validating its potential as a therapeutic target. We are exploring innovative approaches such as PROTAC-based degradation strategies to develop highly selective therapies targeting PFKFB4 for aggressive cancers.
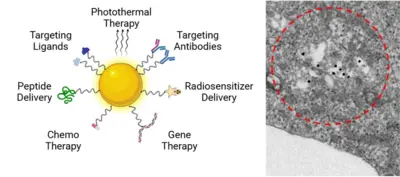
The blood-brain barrier presents a significant obstacle in the therapeutic landscape of glioblastoma. Our research group has identified a promising compound that selectively targets and kills glioblastoma stem-like cells; however, its ability to penetrate the brain is limited. To address this issue, we are developing an innovative gold nanoparticle-based delivery system aimed at enhancing the transport of GSC-targeting drugs across the blood-brain barrier and into the tumor, potentially improving treatment outcomes for this aggressive cancer.
Team
-

Dr. Violaine Goidts
Research Group Leader
-
Dr. Emma Phillips
Postdoc
-
Sized van Enk
PhD student
-
Gabriele Müller
Technical Assistant
Selected Publications
Lokumcu T, Iskar M, Schneider M, Helm D, Klinke G, Schlicker L, Bethke F, Müller G, Richter K, Poschet G, Phillips E, Goidts V.
lhalabi OT, Fletcher MNC, Hielscher T, Kessler T, Lokumcu T, Baumgartner U, Wittmann E, Schlue S, Göttmann M, Rahman S, Hai L, Hansen-Palmus L, Puccio L, Nakano I, Herold-Mende C, Day BW, Wick W, Sahm F, Phillips E, Goidts V.
Phillips E, Balss J, Bethke F, Pusch S, Christen S, Hielscher T, Schnölzer M, Fletcher MNC, Habel A, Tessmer C, Brenner LM, Göttmann M, Capper D, Herold-Mende C, von Deimling A, Fendt SM, Goidts V.
Get in touch with us

