Molecular Hematology/Oncology
- NCT
- Cell and Tumor Biology
- Clinical Cooperation Unit

Prof. Dr. Alwin Krämer
90% of cancer deaths in Germany are caused by metastases, which represent one of the greatest challenges in cancer research. In order to improve treatment outcomes, an adequate understanding of the highly complex metastasis processes and the translation of research findings into effective treatment approaches are required.
Image: @ dkfz, Bianca Kraft,
Image: @ dkfz, Bianca Kraft,
Our Research
Our translational and clinical research focus on Cancer of Unknown Primary (CUP). CUP is a highly aggressive, primary metastatic malignancy, in which only metastases but no primary tumor can be identified. Due to the lack of an identifiable primary tumor, treatment options are limited and patients suffering from CUP experience a dismal prognosis.
Owing to its primary metastatic nature, CUP constitutes a paradigm metastatic disease and therefore enables insights into mechanisms of cancer progression and metastatic seeding in general. Moreover, as the tissue of origin remains elusive, CUP additionally serves as a true tumor-agnostic model disease.
Within this scope, our translational research program aims to unravel the mechanisms underlying cancer metastasis starting with in vitro models and extending into work with primary patient samples (tumor tissue, blood, patient-derived tumoroids), while constantly linking the results to clinical parameters. Our basic science projects center around Chromosomal Instability (CIN) as one of the major mechanisms paving the way towards metastasis. Using genome-wide DNA sequencing, transcriptomics and methylome analysis, we have gained essential insights into the key molecular features of CUP. Based on these findings, and with the ultimate goal of improving treatment options and developing innovative strategies to overcome treatment failure, we conduct large multi-center and international precision medicine clinical trials for newly diagnosed as well as relapsed or refractory treatment settings.
Our efforts have led to the largest randomized phase II trial ever conducted in CUP, which recently and for the first time proved the efficacy of tumor-agnostic, molecularly tailored treatments, constituting a major milestone in the treatment of CUP and a decisive step towards integrating precision oncology into standard medical practice.
Our Projects in detail
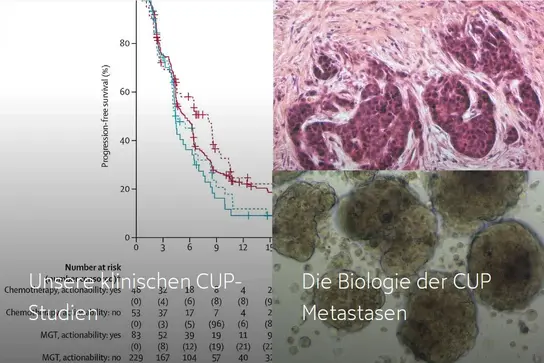
If you would like to find out more, please click on the links below:
Our clinical CUP trials →
The biology of CUP metastases →
Future Outlook
Our future clinical trials focus on the use of antibody-drug-conjugates, a promising approach that has revolutionized the treatment of several cancer entities, in relapsed or refractory CUP syndrome.
Using patient-derived samples acquired within clinical trials, we use multi-omics approaches to molecularly profile and classify CUP tumors, understand CUP biology as well as tumor-host interactions, and predict treatment efficacy based on tumoroid models high-throughput drug screening. Within an integrated approach, and while firmly linking research and clinical practice, we aim to determine CUP tissues of origin and their relevance for treatment results, while enabling molecularly informed therapies, thereby contributing to innovative diagnostic and treatment strategies and improved patient outcomes.
Due to the tumor-agnostic nature of CUP, we will ultimately use the data assembled to transform cancer nomenclature from being based on merely clinicopathological features into a true molecular classification system.
Our Team
As Clinical Cooperation Unit (CCU), we are a joint department of the German Cancer Research Center (DKFZ) and the Department of Internal Medicine V at Heidelberg University Hospital, which is involved in both the research and treatment of CUP.
Our team includes scientists from the fields of human medicine, data science, cell and molecular biology as well as physicians from the University Hospital and the National Center for Tumor Diseases (NCT); they are supported by highly motivated students (PhD, MD students) as well as biological and medical-technical assistants.

Selected Publications
Pouyiourou M, Bochtler T, Pauli C, Moch H, Brobeil A, Pantel K, Stenzinger A, Krämer a
Krämer A, Bochtler T, Pauli C, Shiu KK, Cook N, Janoski de Menezes J, Pazo-Cid RA, Losa F, Robbrecht DGJ, Tomášek J, Arslan C, Özgüroğlu M, Stahl M, Bigot F, Kim SY, Naito Y, Italiano A, Chalabi N, Durán-Pacheco G, Michaud C, Scarato J, Thomas M, Ross JS, Moch H, Mileshkin L
Budczies J, Kazdal D, Menzel M, Beck S, Kluck K, Altbürger C, Schwab C, Allgäuer M, Ahadova A, Kloor M, Schirmacher P, Peters S, Krämer A, Christopoulos P, Stenzinger A
Pouyiourou M*, Kraft BN*, Wohlfromm T, Stahl M, Kubuschok B, Löffler H, Hacker UT, Hübner G, Weiss L, Bitzer M, Ernst T, Schütt P, Hielscher T, Delorme S, Kirchner M, Kazdal D, Ball M, Kluck K, Stenzinger A, Bochtler T, Krämer A
Krämer A, Bochtler T, Pauli C, Baciarello G, Delorme S, Hemminki K, Mileshkin L, Moch H, Oien K, Olivier T, Patrikidou A, Wasan H, Zarkavelis G, Pentheroudakis G, Fizazi K, on behalf of the ESMO Guidelines Committee
Get in touch with us