CCU Molecular Hematology/Oncology - Translational Research
Within the frame of our clinical trials, our translational research program aims to identify new treatment response predictors and to explore the mechanisms responsible for early and aggressive metastatic seeding in CUP. One question has long been the subject of controversy: Is CUP a separate entity that behaves biologically differently than metastases from known primary tumors?
Together with the NCT Tissue Bank Heidelberg, part of the Institute of Pathology at the University Hospital Heidelberg, we collect, characterize and store tumor tissue samples and other bio specimens (blood cells, serum, plasma) from our CUP patients to build up a national CUP biobank.
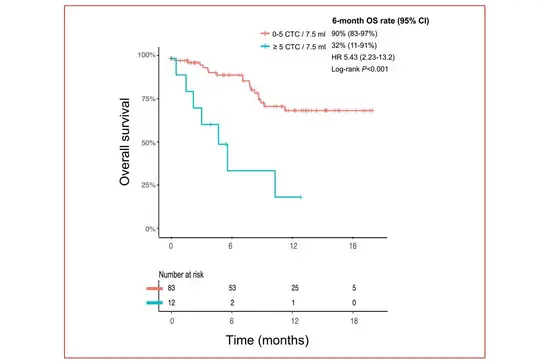
Given the aggressive nature of CUP, reliable early-on monitoring of therapeutic response is instrumental. In a prospective study, we have recently investigated the frequency and prognostic value of circulating tumor cells (CTCs) in 180 blood samples from a cohort of 100 CUP patients. Using a EpCAM-based detection methods, we were able to detect CTCs in 26% of these CUP patients, with the highest frequency of CTC in patients with unfavorable CUP. High CTC levels (≥5 CTCs/7.5 mL) predicted for adverse overall survival compared to negative or low CTC counts. Besides, monitoring CTC levels over time may help to assess treatment response and guide clinical decision-making.
Kaplan-Meier plot showing the overall survival of CUP patients with negative/low CTC frequency (0-5 CTCs/7.5ml) versus patients with high CTC frequency (≥5 CTCs/7.5 ml).
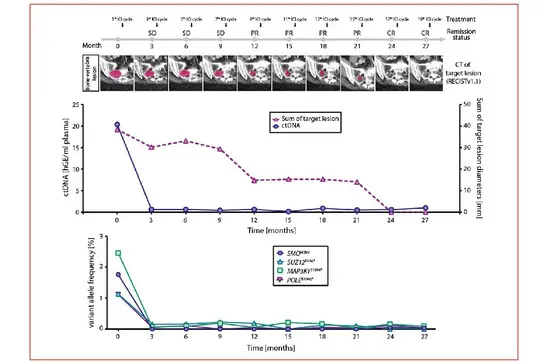
In a second approach, we assessed early on-treatment dynamics of circulating tumor DNA (ctDNA) using combined ultra-deep targeted next-generation sequencing (NGS) of patient-specific hotspot mutations and copy number alteration (CNA) profiling by low-coverage, shallow whole genome sequencing (sWGS) as part of a predefined exploratory analysis during our CheCUP trial. We were able to identify patients deriving long-term treatment benefit irrespective of initial radiologic response.
Representative case of a CUP patient exemplifying the utility of ctDNA analyses in parallel to radiological assessment. Upper graph: Comparison of ctDNA level in serial collected plasma sample with the measured sum of target lesion diameters by RECIST v1.1. Lower graph: Dynamic tracking of VAF from single somatic tumor mutations in ctDNA.
Since driver mutations and gene arrangements can guide treatment individually (precision oncology), a long-standing focus of our group is to identify new pathogenic biomarkers and prognostic factors by comprehensive genomic profiling of our CUP patients.
Ongoing projects
We employ genome-wide DNA, RNA and methylation profiling of large cohorts of primary CUP tumor samples for the development of computational tools for tissue-of-origin identification.
Using multiplexed ion beam imaging (MIBI), a spatial omics strategy that allows for multiplexed in situ imaging of up to 40 antigens, we explore the interplay between tumor, tumor microenvironment and immune response to facilitate a detailed understanding of the mechanisms leading to early and aggressive metastasis on the one hand and absence of a primary tumor on the other hand in patients with CUP.
Using CUP patient-derived tumoroids, we perform in vitro drug screening and functional in-depth characterization of the role of different mutations and chromosomal instability for metastasis formation, clonal evolution and heterogeneity, with CUP syndrome serving as a paradigm disorder for metastatic malignancies.
Project members
-
Dr. Bianca Kraft
Postdoc, PhD, Deputy
-
Dr. Abhishek Kumar
Postdoc, Bioinformatician
-
Maria Pouyiourou
Postdoc, MD
-
Gaia Barberis Canonico
PhD student
-
Maximilian Bieniara
PhD student
-
Elena Kharitonova
MD student
-
Maximilian Haas
MD student
-
Katharina Brobeil
Technician
-
Michael Kirsch
Technician
-
Judith Müller
Technician
Publications (selected)
Tumour mutational burden: clinical utility, challenges and emerging improvements.
Budczies J, Kazdal D, Menzel M, Beck S, Kluck K, Altbürger C, Schwab C, Allgäuer M, Ahadova A, Kloor M, Schirmacher P, Peters S, Krämer A, Christopoulos P, Stenzinger A
Frequency and prognostic value of circulating tumor cells in cancer of unknown primary.
Pouyiourou M, Bochtler T, Coith C, Wikman H, Kraft B, Hielscher T, Stenzinger A, Riethdorf S, Pantel K, Krämer A
Baseline mutational profiles of patients with carcinoma of unknown primary origin enrolled in the CUPISCO study.
Westphalen CB, Federer-Gsponer J, Pauli C, Karapetyan AR, Chalabi N, Durán-Pacheco G, Beringer A, Bochtler T, Cook N, Höglander E, Jin DX, Losa F, Mileshkin L, Moch H, Ross JS, Sokol ES, Tothill RW, Krämer A
Nivolumab and ipilimumab in recurrent or refractory cancer of unknown primary: a phase II trial.
Pouyiourou M*, Kraft BN*, Wohlfromm T, Stahl M, Kubuschok B, Löffler H, Hacker UT, Hübner G, Weiss L, Bitzer M, Ernst T, Schütt P, Hielscher T, Delorme S, Kirchner M, Kazdal D, Ball M, Kluck K, Stenzinger A, Bochtler T, Krämer A
Prognostic impact of copy number alterations and tumor mutational burden in carcinoma of unknown primary.
Bochtler T, Wohlfromm T, Hielscher T, Stichel D, Pouyiourou M, Kraft B, Neumann O, Endris V, von Deimling A, Stenzinger A, Krämer A
Local ablative treatment with surgery and/or radiotherapy in single-site and oligometastatic carcinoma of unknown primary (CUP).
Pouyiourou M, Wohlfromm T, Kraft B, Hielscher T, Stichel D, von Deimling A, Delorme S, Endris V, Neumann O, Stenzinger A, Krämer A, Bochtler T
Comprehensive genomic profiling of carcinoma-of-unknown-primary-origin: retrospective molecular classification considering the CUPISCO study design.
Ross JS, Sokol ES, Moch H, Mileshkin L, Baciarello G, Losa F, Beringer A, Thomas M, Elvin J, Ngo N, Jin DX, Krämer A
Integrated clinicomolecular characterization identifies RAS activation and CDKN2A deletion as independent adverse prognostic factors in cancer of unknown primary.
Bochtler T, Reiling A, Endris V, Hielscher T, Volckmar AL, Neumann O, et al.
Comparative genetic profiling aids diagnosis and clinical decision making in challenging cases of CUP syndrome.
Bochtler T, Endris V, Leichsenring J, Reiling A, Neumann O, Volckmar AL, et al.
RNA-Based Detection of Gene Fusions in Formalin-Fixed and Paraffin-Embedded Solid Cancer Samples.
Kirchner M, Neumann O, Volckmar AL, Stögbauer F, Allgäuer M, Kazdal D, et al.
Germline genetics of cancer of unknown primary (CUP) and its specific subtypes.
Hemminki K, Chen B, Kumar A, Melander O, Manjer J, Hallmans G, et al.
Molecular driver alterations and their clinical relevance in cancer of unknown primary site.
Löffler H, Pfarr N, Kriegsmann M, Endris V, Hielscher T, Lohneis P, et al.
