CCU Molecular Hematology/Oncology - Basic Research
Chromosomal instability (CIN), a central feature of human malignancies, contributes to genetic heterogeneity, clonal evolution and ultimately to metastasis. Accordingly, our group has a long-standing interest in exploring the molecular mechanisms underlying CIN and untangling its role in the metastatic process and the development of CUP as well as metastatic cancer in general.
Centrosomal aberrations, CIN and metastasis
Chromosomal instability (CIN) describes a form of genome instability characterized by high rates of gains and losses of whole chromosomes (numerical CIN) or large chromosome fragments (structural CIN). It is found in over 90% of solid tumors and in blood cancers. Regarding the mechanisms leading to CIN, we are especially interested in the regulation of mitosis and its disturbance in the context of tumorigenesis.
Since centrosomal aberrations have been shown to lead to mitotic aberrations and chromosomal instability, a long-standing focus of our group is centrosome biology and its interconnection to malignant transformation and metastasis. Centrosomes are the major microtubule-organizing centers in mammalian cells and organize the bipolar spindle that partitions chromosomes during mitosis. In normal cells, the centrosomes consist of a pair of centrioles embedded in pericentriolar material.
We have recently generated and in-depth characterized transgenic mice that overexpress the centriole replication protein STIL, in order to gain insights into the role of supernumerary centrosomes in CIN induction and subsequent tumor formation in vivo.
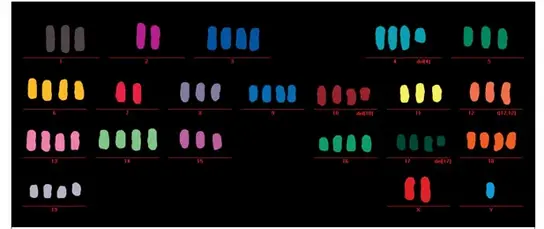
Extra centrosomes induced by STIL overexpression in transgenic mice cause chromosome missegregation and aneuploidy. Shown here is a multiplex fluorescence in situ hybridization (M-FISH) metaphase of a mouse embryonic fibroblast derived from CMV-STIL+/+ mice.
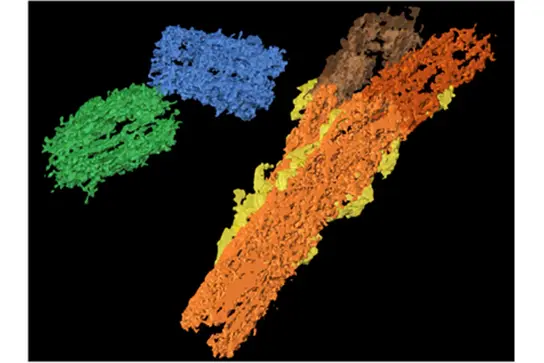
To map the landscape of centrosome aberrations in primary tumor tissues at high resolution, we employ correlative light and electron microscopy (CLEM), focused ion beam scanning electron microscopy (FIB/SEM) and EM tomography with subsequent 3D reconstruction and quantitative analysis. Using these approaches we were able to demonstrate for the first time at nanoscale resolution that centriole over-elongation and the subsequent formation of gross structural centrosome aberrations frequently occur in primary human cells and seem to be associated with cell aging.
Segmented electron tomography image showing an over-elongated centriole (orange) with additional structural abnormalities in a plasma cell from human bone marrow.
Project members
-
Dr. Bianca Kraft
Postdoc, PhD, Deputy
-
Huining Dong
MD student
-
Katharina Brobeil
Technician
-
Michael Kirsch
Technician
-
Judith Müller
Technician
Publications (selected)
STIL overexpression shortens lifespan and reduces tumor formation in mice.
Moussa AT, Cosenza MR, Wohlfromm T, Brobeil K, Hill A, Patrizi A, Müller-Decker K, Holland-Letz T, Jauch A, Kraft B, Krämer A
Single-cell analysis reveals dynamic clonal evolution and targetable phenotypes in complex karyotype AML.
Leppä AM, Grimes K, Jeong H, Huang FY, Delgado AA, Boch T, Waclawiczek A, Jauch A, Renders S, Müller-Tidow C, Karpova D, Sohn M, Grünschläger F, Hasenfeld P, Garagorri EB, Thiel V, Dolnik A, Rodriguez-Martin B, Bullinger L, Mrózek K, Eisfeld AK, Krämer A, Sanders AD, Korbel JO, Trumpp A
High-throughput electron tomography identifies centriole over-elongation as an early event in plasma cell disorders.
Köhrer S, Dittrich T, Schorb M, Weinhold N, Haberbosch I, Bormel M, Pajor G, Goldschmidt H, Müller-Tidow C, Raab MS, John L, Seckinger A, Brobeil A, Dreger P, Tornoczky T, Pajor L, Hegenbart U, Schönland S, Schwab Y, Krämer A
Protocol for high-throughput electron tomography exemplified on centrioles in primary human plasma cells.
Köhrer S, Dittrich T, Schorb M, Haberbosch I, Schwab Y, Krämer A
A high-throughput electron tomography workflow reveals over-elongated centrioles in relapsed-refractory multiple myeloma.
Dittrich T, Köhrer S, Schorb M, Haberbosch I, Börmel M, Goldschmidt H, Müller-Tidow C, Raab MS, Hegenbart U, Schönland SO, Schwab Y, Krämer A
Human SLFN5 and its Xenopus Laevis Ortholog Regulate Entry into Mitosis and Oocyte Meiotic Resumption.
Vit G, Hirth A, Neugebauer N, Kraft B, Sigismondo G, Cazzola A, Tessmer C, Duro J, Krijgsveld J, Hofmann I, Berger M, Klüter H, Niehrs C, Nilsson J, Krämer A
TP53 deficiency permits chromosome abnormalities and karyotype heterogeneity in acute myeloid leukemia.
Cazzola A, Schlegel C, Jansen I, Bochtler T, Jauch A, Krämer A
SMC3 protein levels impact on karyotype and outcome in acute myeloid leukemia.
Kraft B, Lombard J, Kirsch M, Wuchter P, Bugert P, Hielscher T, et al.
Micronucleus formation in human cancer cells is biased by chromosome size.
Bochtler T, Kartal-Kaess M, Granzow M, Hielscher T, Cosenza MR, Herold-Mende C, et al.
Cytogenetic intraclonal heterogeneity of plasma cell dyscrasia in AL amyloidosis as compared with multiple myeloma.
Bochtler T, Merz M, Hielscher T, Granzow M, Hoffmann K, Krämer A, et al.
Asymmetric Centriole Numbers at Spindle Poles Cause Chromosome Missegregation in Cancer.
Cosenza MR, Cazzola A, Rossberg A, Schieber NL, Konotop G, Bausch E, et al.
Marker chromosomes can arise from chromothripsis and predict adverse prognosis in acute myeloid leukemia.
Bochtler T, Granzow M, Stölzel F, Kunz C, Mohr B, Kartal-Kaess M, et al.
Pharmacological Inhibition of Centrosome Clustering by Slingshot-Mediated Cofilin Activation and Actin Cortex Destabilization.
Konotop G, Bausch E, Nagai T, Turchinovich A, Becker N, Benner A, et al.
Karyotype complexity and prognosis in acute myeloid leukemia.
Stölzel F, Mohr B, Kramer M, Oelschlägel U, Bochtler T, Berdel WE, et al.
Cep63 recruits cdk1 to the centrosome-letter.
Alsara M, Löffler H, Fechter A, Bartek J, Krämer A
Role of chromosomal aberrations in clonal diversity and progression of acute myeloid leukemia.
Bochtler T, Fröhling S, Krämer A
Clonal heterogeneity as detected by metaphase karyotyping is an indicator of poor prognosis in acute myeloid leukemia.
Bochtler T, Stölzel F, Heilig CE, Kunz C, Mohr B, Jauch A, et al.
EGF-induced centrosome separation promotes mitotic progression and cell survival.
Mardin BR, Isokane M, Cosenza MR, Krämer A, Ellenberg J, Fry AM, Schiebel E
DNA damage-induced centrosome amplification occurs via excessive formation of centriolar satellites.
Löffler H, Fechter A, Liu FY, Poppelreuther S, Krämer A
STIL is required for centriole duplication in human cells.
Vulprecht J, David A, Tibelius A, Castiel A, Konotop G, Liu FY, et al.
GF-15, a novel inhibitor of centrosomal clustering, suppresses tumor cell growth in vitro and in vivo.
Raab MS, Breitkreutz I, Anderhub S, Rønnest MH, Leber B, Larsen TO, et al.
Genomic instability and myelodysplasia with monosomy 7 consequent to EVI1 activation after gene therapy for chronic granulomatous disease.
Stein S, Ott MG, Schultze-Strasser S, Jauch A, Burwinkel B, Kinner A, et al.
Proteins required for centrosome clustering in cancer cells.
Leber B, Maier B, Fuchs F, Chi J, Riffel P, Anderhub S, et al.
Microcephalin and pericentrin regulate mitotic entry via centrosome-associated Chk1.
Microcephalin and pericentrin regulate mitotic entry via centrosome-associated Chk1.
Identification of griseofulvin as an inhibitor of centrosomal clustering in a phenotype-based screen.
Rebacz B, Larsen TO, Clausen MH, Rønnest MH, Löffler H, Ho AD, Krämer A
DNA damage response as a candidate anti-cancer barrier in early human tumorigenesis.
Bartkova J, Horejsí Z, Koed K, Krämer A, Tort F, Zieger K, et al.
Centrosome-associated Chk1 prevents premature activation of cyclin-B-Cdk1 kinase.
Krämer A, Mailand N, Lukas C, Syljuåsen RG, Wilkinson C, Nigg EA, et al.
Centrosome aberrations in acute myeloid leukemia are correlated with cytogenetic risk profile.
Neben K, Giesecke C, Schweizer S, Ho AD, Krämer A
