Molecular Neurogenetics
- Cell and Tumor Biology

Dr. Haikun Liu
Head of Division
The Division of Molecular Neurogenetics focuses on stem cell biology and central nervous system cancers. Our primary objective is to decipher the fundamental mechanisms driving brain tumor plasticity and to develop innovative therapies that improve glioblastoma treatment.
Image: Organoid @ DKFZ,
Image: Organoid @ DKFZ,
Our Research
In the Division of Molecular Neurogenetics, our research centers on uncovering the molecular mechanisms that drive brain development and pathology, with a special focus on brain cancers. Over the past several years, we have identified key regulators of brain tumor stem cells, including TLX and GPD1. Notably, our work demonstrated that knocking out these factors confers a survival benefit in vivo (Cell Stem Cell, 2014), a finding recognized as one of the year’s best papers and highlighted in Nature Reviews Cancer.
More recently, we identified GPD1 as the first known dormant brain tumor stem cell marker and essential regulator (Cell Stem Cell, 2019), providing critical insight into an emerging research area of cancer dormancy across various tumor types. Additionally, our studies have illuminated the crucial role of the CHARGE syndrome protein CHD7 in governing cell differentiation, a discovery that underpins a fundamental understanding of CHARGE syndrome (Cell Stem Cell, 2013; Nature Communications, 2017).
Future Outlook:
Our current efforts involve generating sophisticated human brain organoid and brain tumor organoid models that closely replicate the genetic and phenotypic features of brain tumors(NPJ Precis Oncol. 2024, Cell Stem Cell, 2025). Leveraging state-of-the-art single-cell technology, CRISPR/Cas9 genome editing and artificial intelligence, we aim to dissect the molecular and cellular heterogeneity in human glioblastoma, with a strong emphasis on dormant cancer stem cells. Ultimately, we aim to translate our fundamental findings into clinical settings, working toward effective treatments for glioblastoma, one of the most lethal human cancers.
Projects
- Targeting cancer stem cells
- Modeling human cancer using LEGO organoid
- Predictive Patient Derived Models for Personalized Neurooncology
- Molecular principles in neural stem cells, neurogenesis and brain diseases
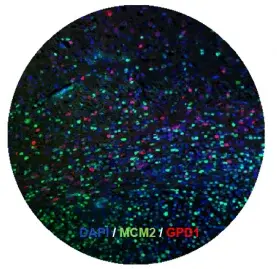
Our previously discoveries demonstrated that cancer stem cells are the right target in glioblastoma (2014, Cell Stem Cell). We recently identified the first dormant brain tumor stem cells (2019, Cell Stem Cell), which are resistant to conventional therapies. Along this line, we also identified crucial regulators of cancer stem cell dormancy. Currently we are developing several compounds aiming eliminating brain tumor stem cells, which will be further tested in clinical trials. In this direction, we are particularly interested in targeting metabolic pathways regulating tumor cell metabolism.
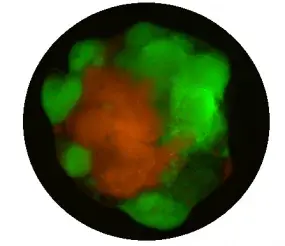
To systematically model the genetic spectrum affecting human glioblastoma (GBM), we employ human induced pluripotent stem cell (iPSC)-derived organoids to elucidate the genetic heterogeneity within GBM. Utilizing CRISPR/Cas9, we generated a comprehensive spectrum of mutation combinations (Pten, Trp53, Cdkn2a, Cdkn2b, Nf1, Rb1, Egfr, Tert, etc.), which are the most frequently mutated genes in human GBM, in human iPSCs. Subsequently, we induced GBM organoids from these cells, which we refer to as LEGO: Laboratory Engineered Glioma Organoid (npj Preci Onco, 2024). These organoids effectively recapitulate key molecular features of human GBM.
In the LEGO2.0 model, we further enhance its fidelity by incorporating barcodes, immune cells, and blood vessels. This comprehensive model enables us to systematically dissect the interaction between genetic heterogeneity and functional heterogeneity within GBM. Additionally, drug screening is underway to establish a genotype-based drug reference for personalized treatment of GBM
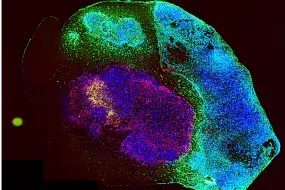
Tumor organoids are pivotal tools in cancer research, yet current models face limitations that hinder their application in predicting therapeutic responses. To address this challenge, we have developed a sophisticated culture system (IPTO, individualized patient tumor organoid, Cell Stem Cell, 2025) that accurately simulates the cellular and molecular pathology of human brain tumors. Patient-derived tumor explants were cultured within induced pluripotent stem cell (iPSC)-derived cerebral organoids, enabling the cultivation of a diverse range of human tumors within the central nervous system (CNS), including adult, pediatric, and metastatic brain cancers. Histopathological, genomic, epigenomic, and single-cell RNA sequencing (scRNA-seq) analyses revealed that the IPTO model accurately recapitulates cellular heterogeneity and molecular features of the original tumors.
Notably, we demonstrated that the IPTO model accurately predicts patient-specific drug responses, including resistance mechanisms, in a prospective patient cohort. Collectively, the IPTO model represents a significant advancement in preclinical modeling of human cancers, paving the way for personalized cancer therapy. The IPTO model is also utilized for target discovery, validation, and T cell-based cancer therapies. The high-quality drug response data generated from the IPTO models are being employed to train predictive generative AI models for patient response prediction.

We generated a series of mouse models and human organoid models that allow us to precisely manipulate gene expression in neural stem cells and brain tumor stem cells. In this project, we are dedicated to unravel how the dynamic transcriptional regulations are involved in regulating the identity, activity and fate of NSCs. We are systematically analyzing the role of essential transcription factors and chromatin regulators in neurogenesis in order to generalize the molecular principles of cell identity determination. In particular we are interested in studying transcription regulators like Nr2e1 and Chd7 which are mutated in human neurological disorders and brain cancers.
Our Team
- Show profile

Dr. Haikun Liu
Head of Division
-

Heike Alter
Technician
-

Yuling Chen
PhD Student
-

Jin Chen
PhD Student
-

Claudia Guevara Lanero
Master Student
-

Yassin Harim
PhD Student
-

Yixuan Hu
-
Sophia Kornherr
Master Student
-

Xian Li
PhD Student
-

Xiaoyu Ma
PhD Student
-

Gabriele Meyer
Foreign-language Secretary
-

Yiwei Qi
-

Miguel Sanchez Oliveros
-

Nadja Stöffler
Technician
-

Dr. Weili Tian
PostDoc
-

Cara Sophie Timmerhoff
-

Angela Valentino
-
Beatrice Valerie Wagner
-

Qing Zheng
Visiting PhD Student
-

Yue Zhuo
PhD Student
Selected Publications
Peng T, Ma X, Hua W, Wang C, Chu Y, Sun M, Fermi V, Hamelmann S, Lindner K, Shao C, Zaman J, Tian W, Zhuo Y, Harim Y, Stöffler N, Hammann L, Xiao Q, Jin X, Warta R, Lotsch C, Zhuang X, Feng Y, Fu M, Zhang X, Zhang J, Xu H, Qiu F, Xie L, Zhang Y, Zhu W, Du Z, Salgueiro L, Schneider M, Eichhorn F, Lefevre A, Pusch S, Grinevich V, Ratliff M, Loges S, Bunse L, Sahm F, Xiang Y, Unterberg A, von Deimling A, Platten M, Herold-Mende C, Wu Y, Liu HK*, Mao Y. (* Lead Contact)
Wang C, Sun M, Shao C, Schlicker L, Zhuo Y, Harim Y, Peng T, Tian W, Stöffler N, Schneider M, Helm D, Chu Y, Fu B, Jin X, Mallm JP, Mall M, Wu Y, Schulze A, Liu HK.
Venkataramani V, Tanev DI, Strahle C, Studier-Fischer A, Fankhauser L, Kessler T, Körber C, Kardorff M, Ratliff M, Xie R, Horstmann H, Messer M, Paik SP, Knabbe J, Sahm F, Kurz FT, Acikgöz AA, Herrmannsdörfer F, Agarwal A, Bergles DE, Chalmers A, Miletic H, Turcan S, Mawrin C, Hänggi D, Liu HK, Wick W, Winkler F, Kuner T.
Rusu P, Shao C, Neuerburg A, Acikgöz AA, Wu Y, Zou P, Phapale P, Shankar TS, Döring K, Dettling S, Körkel-Qu H, Bekki G, Costa B, Guo T, Friesen O, Schlotter M, Heikenwalder M, Tschaharganeh DF, Bukau B, Kramer G, Angel P, Herold-Mende C, Radlwimmer B, Liu HK.
All Publications
Selected Awards and Prizes
- ERC (European Research Council) Consolidator Award for Hai-Kun. DKFZ Press Release
- Hai-Kun receives the Chica and Heinz Schaller research award 2015 from CHS Foundation (Chica and Heinz Schaller Foundation). DKFZ Press Release
- Hai-Kun received the first DKFZ Alumni award for international scientists.
- Hai-Kun was selected to join the EMBO Young Investigator Program in 2014, which represents some of the best young group leaders contributing to research in Europe and beyond.
- ISSCR travel award for Zhe
- Best photo at the picture contest of the DKFZ PhD students for Gözde
- ISSCR travel award for Weijun
- Helmholtz Young Investigator Award for Hai-Kun, funding grant of the lab
Funding

Günther Schütz Prize and Symposium
Get in touch with us









