Molecular Thoracic Oncology
- Functional and Structural Genomics

Prof. Dr. Rocio Sotillo Roman
Group Leader
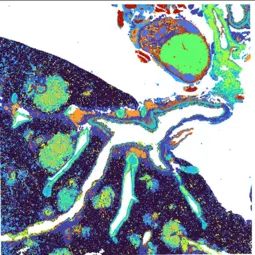
Our Research

Our research is focused on understanding the genetic and molecular basis of lung and breast cancers by studying various oncogenic drivers and specific genetic alterations that drive cancer progression. Using advanced genetically engineered mouse models, we examine how distinct gene expression profiles influence tumor aggressiveness and responsiveness to treatment. A key part of our research involves identifying biomarkers predictive of poor therapeutic outcomes, aiming to develop more precise cancer therapies.
Methods and technologies
Our research leverage several cutting-edge technologies:
- Genetically engineered mouse models to closely mimic human cancer.
- CRISPR/Cas9 technology for precise genetic modifications to study the impact of specific genes on cancer.
- Lineage tracing, this technique traces the evolution and differentiation of cancer cells, offering insights into tumor development.
- Epigenome and single-cell transcriptome analysis, help us explore gene expression and regulatory changes at the single-cell level, enhancing our understanding of cancer cell behavior.
- 3D in vitro culture systems that allow us to study cancer growth in environments that closely mimic physiological conditions.
Goals and societal relevance
The overarching goal of our research is to advance our understanding of cancer biology to discover new therapeutic targets and strategies. By dissecting the molecular mechanisms of tumor progression and genetic instability, we strive to develop more effective treatments. Our efforts support the German Cancer Research Centre’s mission to address major challenges in cancer research, particularly in identifying and overcoming resistance mechanisms to improve patient outcomes.
Projects

Deciphering the Genetic Landscape of Non-Small Cell Lung Cancer: Fusion Variants and Therapeutic Opportunities
Non-small cell lung cancer (NSCLC) is one of the most common and deadly cancers worldwide. A significant driver of NSCLC is the fusion between the genes anaplastic lymphoma kinase (ALK) and echinoderm microtubule-associated protein-like 4 (EML4). To date, approximately 15 distinct EML4-ALK fusion variants have been identified, each with unique characteristics affecting their stability, genomic localization, responsiveness to targeted therapies, and implications for prognosis.
Our research is dedicated to unraveling the intricate mechanisms that drive development and progression of these fusion variants. We are particularly interested in how co-occurring genetic alterations, such as the loss of TP53, influence the behavior of these variants and identify new therapeutic targets. Beyond EML4-ALK fusions, our team is exploring other chromosomal rearrangements that contribute to NSCLC pathogenesis, including the effects of Y-chromosome loss, and the role of specific epigenetic regulators such as Kdm5d and Uty in the disease.
To probe these genetic intricacies, we leverage CRISPR/Cas9 gene editing to generate genetically engineered mouse models, enabling the inducible expression of specific gene fusions and the targeted inactivation of genes associated with NSCLC. Our approach integrates cutting-edge techniques, including tailored in vivo tumor models that mimic human disease, organoids derived from mice and patients, and sequential gene perturbation assays conducted in vivo. This comprehensive methodology not only replicates the histopathological and molecular characteristics observed in human lung cancer but also provides a powerful platform for dissecting NSCLC's molecular drivers and developing groundbreaking treatment options.
Navigating the Evolutionary Dynamics of Her2+ Breast Cancer
Her2+ breast cancer is its known for its dynamic nature, constantly evolving and adapting in response to therapies and immune challenges. Our research focuses on understanding the molecular and genetic mechanisms that drive this evolution, utilizing advanced mouse models to pinpoint new vulnerabilities to improve therapeutic outcomes.
Our previous work has shed light on how chromosomal instability (CIN) contributes to breast cancer progression. We discovered that PLK1 overexpression promotes immune suppression through senescence-associated secretory phenotypes (SASPs) and NF-κβ signaling. Additionally, our studies on FOXM1 highlight its crucial role in helping aneuploid tumor cells manage mitotic stress, marking it as a promising target for therapy.
Recently, our focus has shifted to APOBEC3B (A3B), a DNA-editing enzyme that introduces mutations, thus driving tumor evolution. Leveraging a unique Her2-A3B inducible mouse model, we aim to unravel how A3B influences the initiation and progression of Her2+ breast cancer.
By integrating studies on chromosomal instability, mutational processes, and tumor microenvironment, we are piecing together the intricate puzzle of breast cancer evolution. Our comprehensive approach using various mouse models is guiding us toward more effective therapeutic strategies, offering hope against this highly adaptive disease.
Lab pictures
Team
- Show profile

Prof. Dr. Rocio Sotillo Roman
Group Leader
- Show profile

Maria Bandeira Ferreira Ramos
PhD student
- Show profile

Maria Capone
PhD student
- Show profile

Maria Teresa Castillo Alvarez
PhD student
-

Hilary Ann Davies-Rück
Administrative assistant
- Show profile

Can Gürkaslar
Lab technician
-
Janina Hattemer
Visiting student
- Show profile

Luisa Kinas
Master student
- Show profile

Amelie Mahr
PhD student
- Show profile

Mulham Najajreh
PhD student
- Show profile

Lukas Otto
Master student
- Show profile

Francisco Javier Rios Sola
Erasmus Student
- Show profile

Dr. Kalman Somogyi
Senior scientist
Selected Publications
EML4-ALK variant-specific genetic interactions shape lung tumorigenesis.
Diaz-Jimenez, A., Shuldiner, E. G., Somogyi, K., Gonzalez, O., Akkas, F., Murray, C. W., Andrejka, L., Tsai, M. K., Brors, B., Sivakumar, S., Sisoudiya, S. D., Sokol, E. S., Petrov, D. A., Winslow, M. M., & Sotillo, R
Diaz-Jimenez A, Ramos M, Helm B, Chocarro S, Frey DL, Agrawal S, Somogyi K, Klingmüller U, Lu J, Sotillo R.
Alonso de la Vega A, Temiz NA, Tasakis R, Somogyi K, Salgueiro L, Zimmer E, Ramos M, Diaz-Jimenez A, Chocarro S, Fernández-Vaquero M, Stefanovska B, Reuveni E, Ben-David U, Stenzinger A, Poth T, Heikenwälder M, Papavasiliou N, Harris RS, Sotillo R
All Publications
Monfort-Vengut A, Sanz-Gómez N, Ballesteros-Sánchez S, Ortigosa B, Cambón A, Ramos M, Lorenzo ÁM, Escribano-Cebrián M, Rosa-Rosa JM, Martínez-López J, Sánchez-Prieto R, Sotillo R, de Cárcer G.
de Jaime-Soguero A, Hattemer J, Bufe A, Haas A, van den Berg J, van Batenburg V, Das B, di Marco B, Androulaki S, Böhly N, Landry JJM, Schoell B, Rosa VS, Villacorta L, Baskan Y, Trapp M, Benes V, Chabes A, Shahbazi M, Jauch A, Engel U, Patrizi A, Sotillo R, van Oudenaarden A, Bageritz J, Alfonso J, Bastians H, Acebrón SP.
Chronic spindle assembly checkpoint activation causes myelosuppression and gastrointestinal atrophy.
Karbon G, Schuler F, Braun VZ, Eichin F, Haschka M, Drach M, Sotillo R, Geley S, Spierings DC, Tijhuis AE, Foijer F, Villunger A.
Horvat NK, Chocarro S, Marques O, Bauer TA, Qiu R, Diaz-Jimenez A, Helm B, Chen Y, Sawall S, Sparla R, Su L, Klingmüller U, Barz M, Hentze MW, Sotillo R, Muckenthaler MU.
Diaz-Jimenez A, Ramos M, Helm B, Chocarro S, Frey DL, Agrawal S, Somogyi K, Klingmüller U, Lu J, Sotillo R.
Cai L, Gao Y, DeBerardinis RJ, Acquaah-Mensah G, Aidinis V, Beane JE, Biswal S, Chen T, Concepcion-Crisol CP, Grüner BM, Jia D, Jones R, Kurie JM, Lee MG, Lindahl P, Lissanu Y, Lorz Lopez MC, Martinelli R, Mazur PK, Mazzilli SA, Mii S, Moll H, Moorehead R, Morrisey EE, Ng SR, Oser MG, Pandiri AR, Powell CA, Ramadori G, Santos Lafuente M, Snyder E, Sotillo R, Su KY, Taki T, Taparra K, Xia Y, van Veen E, Winslow MM, Xiao G, Rudin CM, Oliver TG, Xie Y, Minna JD.
Alonso de la Vega A, Temiz NA, Tasakis R, Somogyi K, Salgueiro L, Zimmer E, Ramos M, Diaz-Jimenez A, Chocarro S, Fernández-Vaquero M, Stefanovska B, Reuveni E, Ben-David U, Stenzinger A, Poth T, Heikenwälder M, Papavasiliou N, Harris RS, Sotillo R.
Kandala S, Ramos M, Voith von Voithenberg L, Diaz-Jimenez A, Chocarro S, Keding J, Brors B, Imbusch CD, Sotillo R.
Durfee C, Temiz NA, Levin-Klein R, Argyris PP, Alsøe L, Carracedo S, Alonso de la Vega A, Proehl J, Holzhauer AM, Seeman ZJ, Liu X, Lin YT, Vogel RI, Sotillo R, Nilsen H, Harris RS.
Raach B, Bundgaard N, Haase MJ, Starruß J, Sotillo R, Stanifer ML, Graw F.
Pan F, Chocarro S, Ramos M, Chen Y, Alonso de la Vega A, Somogyi K, Sotillo R.
Durfee C, Temiz NA, Levin-Klein R, Argyris PP, Alsøe L, Carracedo S, de la Vega AA, Proehl J, Holzhauer AM, Seeman ZJ, Lin YT, Vogel RI, Sotillo R, Nilsen H, Harris RS.
Chen Y, Toth R, Chocarro S, Weichenhan D, Hey J, Lutsik P, Sawall S, Stathopoulos GT, Plass C, Sotillo R.
Salgueiro L, Kummer S, Sonntag-Buck V, Weiß A, Schneider MA, Kräusslich HG, Sotillo R.
Lamort AS, Kaiser JC, Pepe MAA, Lilis I, Ntaliarda G, Somogyi K, Spella M, Behrend SJ, Giotopoulou GA, Kujawa W, Lindner M, Koch I, Hatz RA, Behr J, Sotillo R, Schamberger AC, Stathopoulos GT.
Alikhanyan K, Chen Y, Somogyi K, Kraut S, Sotillo R.
Bauer TA, Horvat NK, Marques O, Chocarro S, Mertens C, Colucci S, Schmitt S, Carrella LM, Morsbach S, Koynov K, Fenaroli F, Blümler P, Jung M, Sotillo R, Hentze MW, Muckenthaler MU, Barz M.
Rubio T, Weyershaeuser J, Montero MG, Hoffmann A, Lujan P, Jechlinger M, Sotillo R, Köhn M.
Mayakonda A, Schönung M, Hey J, Batra RN, Feuerstein-Akgoz C, Köhler K, Lipka DB, Sotillo R, Plass C, Lutsik P, Toth R.
Alikhanyan K, Chen Y, Kraut S, Sotillo R.
Salgueiro L, Buccitelli C, Rowald K, Somogyi K, Kandala S, Korbel JO, Sotillo R.
Raimondi F, Inoue A, Kadji FMN, Shuai N, Gonzalez JC, Singh G, de la Vega AA, Sotillo R, Fischer B, Aoki J, Gutkind JS, Russell RB.
Spella M, Lilis I, Pepe MA, Chen Y, Armaka M, Lamort AS, Zazara DE, Roumelioti F, Vreka M, Kanellakis NI, Wagner DE, Giannou AD, Armenis V, Arendt KA, Klotz LV, Toumpanakis D, Karavana V, Zakynthinos SG, Giopanou I, Marazioti A, Aidinis V, Sotillo R, Stathopoulos GT.
Christopoulos P, Kirchner M, Bozorgmehr F, Endris V, Elsayed M, Budczies J, Ristau J, Penzel R, Herth FJ, Heussel CP, Eichhorn M, Muley T, Meister M, Fischer JR, Rieken S, Lasitschka F, Bischoff H, Sotillo R, Schirmacher P, Thomas M, Stenzinger A.
de Cárcer G, Venkateswaran SV, Salgueiro L, El Bakkali A, Somogyi K, Rowald K, Montañés P, Sanclemente M, Escobar B, de Martino A, McGranahan N, Malumbres M, Sotillo R.
Havas KM, Milchevskaya V, Radic K, Alladin A, Kafkia E, Garcia M, Stolte J, Klaus B, Rotmensz N, Gibson TJ, Burwinkel B, Schneeweiss A, Pruneri G, Patil KR, Sotillo R, Jechlinger M.
Domingues PH, Nanduri LSY, Seget K, Venkateswaran SV, Agorku D, Viganó C, von Schubert C, Nigg EA, Swanton C, Sotillo R, Bosio A, Storchová Z, Hardt O.
Buccitelli C, Salgueiro L, Rowald K, Sotillo R, Mardin BR, Korbel JO.
Weischenfeldt J, Dubash T, Drainas AP, Mardin BR, Chen Y, Stütz AM, Waszak SM, Bosco G, Halvorsen AR, Raeder B, Efthymiopoulos T, Erkek S, Siegl C, Brenner H, Brustugun OT, Dieter SM, Northcott PA, Petersen I, Pfister SM, Schneider M, Solberg SK, Thunissen E, Weichert W, Zichner T, Thomas R, Peifer M, Helland A, Ball CR, Jechlinger M, Sotillo R, Glimm H, Korbel JO.
Rowald K, Mantovan M, Passos J, Buccitelli C, Mardin BR, Korbel JO, Jechlinger M, Sotillo R.
Schvartzman JM, Duijf PH, Sotillo R, Coker C, Benezra R.
Aneiros-Fernandez J, Arias-Santiago S, Sotillo R, Menjon-Beltran S, Concha A.
Sotillo R, Schvartzman JM, Socci ND, Benezra R.
Schvartzman JM, Sotillo R, Benezra R.
Sotillo R, Schvartzman JM, Benezra R.
Diaz-Rodríguez E, Sotillo R, Schvartzman JM, Benezra R.
Niault T, Hached K, Sotillo R, Sorger PK, Maro B, Benezra R, Wassmann K.
Sotillo R, Hernando E, Díaz-Rodríguez E, Teruya-Feldstein J, Cordón-Cardo C, Lowe SW, Benezra R.
Barbacid M, Ortega S, Sotillo R, Odajima J, Martín A, Santamaría D, Dubus P, Malumbres M.
Sotillo R, Renner O, Dubus P, Ruiz-Cabello J, Martín-Caballero J, Barbacid M, Carnero A, Malumbres M.
Malumbres M, Sotillo R, Santamaría D, Galán J, Cerezo A, Ortega S, Dubus P, Barbacid M.
Ortega S, Prieto I, Odajima J, Martín A, Dubus P, Sotillo R, Barbero JL, Malumbres M, Barbacid M.
Martín J, Hunt SL, Dubus P, Sotillo R, Néhmé-Pélluard F, Magnuson MA, Parlow AF, Malumbres M, Ortega S, Barbacid M.
Malumbres M, Hunt SL, Sotillo R, Martín J, Odajima J, Martín A, Dubus P, Ortega S, Barbacid M.
Sotillo R, Dubus P, Martín J, de la Cueva E, Ortega S, Malumbres M, Barbacid M.
Sotillo R, García JF, Ortega S, Martin J, Dubus P, Barbacid M, Malumbres M.
Latres E, Malumbres M, Sotillo R, Martín J, Ortega S, Martín-Caballero J, Flores JM, Cordón-Cardo C, Barbacid M.
Get in touch with us







