Metabolomics
The Metabolomics Core Technology Platform (MCTP) is a DFG-registered (RI_00492) core facility of the Heidelberg University and officially collaborates with DKFZ. It is part of the Centre for Organismal Studies (COS) and serves as an official core facility within the CellNetworks Core Technology Platform (CCTP).

Project Requests
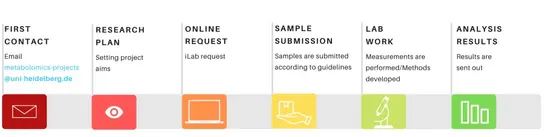
All researchers from DKFZ are welcome to request help or place orders for metabolite analyses.
Targeted metabolomics focuses on the precise analysis of predefined metabolites across all biological matrices. These assays enable absolute quantification by utilizing external or internal standards.
Typically, targeted assays are applied to low-abundance compounds that may not be detected in untargeted explorative analyses.
Our services include:
Metabolite extraction, separation, and detection
Basic bioinformatics analysis of the acquired data
If you require targeted analysis of metabolites beyond those listed, please contact MCTP for customized solutions.
Download Documents
Untargeted metabolomics enables a comprehensive screening of all detectable compounds using validated chromatography and mass spectrometry methods.
Depending on the sample type and analytical platform, 10–30% of detected peaks can be identified based on retention times and/or mass spectra, using in-house, commercial, and publicly available databases. This approach also detects structurally unknown compounds, supporting explorative analyses.
Quantitative data are typically reported as:
Fold changes between sample groups
Relative peak intensities, normalized to quality control samples or internal standards
Download Documents
Large-scale metabolome profiling is available for biofluids, cells, and tissue samples from various origins, utilizing the validated Biocrates MxP® Quant 500 and Quant 500 XL kits.
These assays enable the absolute quantification of up to 630 or more than 1,000 metabolites, covering a broad range of key metabolite and lipid classes.
This technique is particularly well-suited for:
Studies involving human biofluids or tissue samples
Large-scale cohort studies requiring high-throughput and standardized metabolomic analysis
Download Documents
13C metabolic tracing is a powerful technique for investigating enzymatic reaction rates in cells or in vivo systems by tracking the incorporation of heavy-labeled tracer molecules (e.g., ¹³C-glucose or ¹³C-glutamine).
Using advanced GC-TOF and GC-MS/MS analysis, we can determine ¹³C incorporation into up to 50 key metabolic compound classes, including amino acids, glycolysis and TCA cycle intermediates and certain nucleotides
This approach provides crucial insights into metabolic flux and pathway activity in diverse biological systems.
Download Documents
--- UNDER DEVELOPMENT ---
We are currently establishing an MS-based Imaging Platform for spatially resolved 2D and 3D metabolomic profiling. This platform, generously funded by the Health + Life Science Alliance Heidelberg Mannheim, is administered by MCTP and jointly coordinated by C. Opitz (DKFZ), C. Hopf (HS Mannheim), and G. Poschet (MCTP).
General Information
- Prokaryotes
- Yeast
- Cell cultures
- Tissue, e.g. human, animal, plant
- Body fluids, e.g. blood, urine, CSF
- Supernatants, e.g. from cultures
- Further matrcies can be established on request
Before sample submission, please:
- Follow the steps outlined in our sample submission checklist.
- Carefully read the general guidelines for metabolite analyses, including specific requirements.
- Review our End-User License Agreement (EULA) for detailed information on MCTP services, as well as user rights and responsibilities. New customers must provide a signed copy of the EULA when submitting their first sample set.
Core facility services, including system usage and consumables, are directly charged to users. For detailed pricing, please contact MCTP or log in to the online booking system to view available service options.
The MCTP utilizes iLab Solutions, an online platform that streamlines service requests, ordering, and billing for core facility users.
HOURS
MON - 10:30 – 11:30
TUE - 9:00 – 11:00
WED - 13:30 – 15:00
THU - 9:00 – 11:00
Other times upon prior consultation via Email
Shipping address for shipment of samples via post:
MCTP
Centre for Organismal Studies
Universität Heidelberg
Im Neuenheimer Feld 360
69120 Heidelberg
Germany
Instrumentation
High-sensitivity triple-quad UPLC-MS/MS instruments for quantitative targeted assays (Sciex QTRAP 6500+)
High-resolution, high mass accuracy UPLC-QTOF with ion-mobility separation, mainly for untargeted metabolomics (Waters VION IMS-QTOF, Sciex ZenoTOF)
GC-TOF and GC-MS/MS instrument for central metabolism screening, untargeted metabolomics and 13C tracing studies (Leco BT-TOF, Shimadzu GCMS-TQ8040 NX)
GC-MS for analysis of free fatty acids (Shimadzu GCMS-QP2010)
Various LC systems coupled to optical and special detectors & automated pipetting systems (Eppendorf EpMotion)
Our equipment for spatial metabolomics consists of a timsTOF FLEX MALDI-2 system, a laser microdissection system and a cryomicrotome (located at BioQuant)
Team & Contact
Team
- Scientific Director Prof. Dr. Rüdiger Hell
- Managing Director Dr. Gernot Poschet
- Scientist for DKFZ Projects Erwin Kupczyk
- Scientific Staff Dr. Michael Büttner
- Technical Assistant Nina Kunze-Rohrbach
- Technical Assistant Michael Schulz
- Administration Birgit Maresch
Contact
Where to find us:
Im Neuenheimer Feld 360 (in the Botanical Garden)
Room -118
D- 69120 Heidelberg
Who to contact:
Dr. Gernot Poschet
Email: gernot.poschet(at)cos.uni-heidelberg.de
Erwin Kupczyk
Email: erwin.kupczyk(at)cos.uni-heidelberg.de
Selected Publications
Couble JE, Vignolini T, Doré G, Mussgnug S, Lombard B, Richard M, Loew D, Büttner M, Dueñas-Sánchez R, Poschet G, Bryant JM, Baumgarten S.
Seifert S, Yanqui-Rivera F, Kühn T, Høyland LE, Borteçen T, Agranovich B, Eckhardt LE, Herdt M, Henneberg A, Panitz V, Hayat F, Migaud M, Poschet G, Abramovich I, Gottlieb E, Krijgsveld J, Ziegler M, Prentzell MT, Opitz CA.
Söder L, Baeza-Lehnert F, Khodaie B, Elgez A, Noack L, Lewen A, Hallermann S, Poschet G, Borges K, Kann O.
Sulaj A, Nguyen PBH, Poschet G, Kliemank E, Fleming T, Henke L, Neibig W, Kopf S, Hell R, Longo VD, Herzig S, Nawroth PP, Menden MP, Szendroedi J.
Figlia G, Müller S, Garcia-Cortizo F, Neff M, Klinke G, Poschet G, Teleman AA.
Falkenstein K, Hoeren L, Kikul F, Poschet G, Lüchtenborg C, Brecht IB, Falb R, Gauck D, Haack T, Hecker A, Himmelreich N, Okun JG, Brügger B, Thiel C.
Liu H, Tang Y, Singh A, Vong J, Cordero J, Mathes A, Gao R, Jia Y, Garvalov BK, Acker T, Poschet G, Hell R, Schneider MA, Heineke J, Wieland T, Barreto G, Cerwenka A, Potente M, Bibli SI, Savai R, Dobreva G.
Xu F, Jansakun C, Li G, Biswas U, Poschet G, Staffer S, Tuma-Kellner S, Nakchbandi I, Merle U, Chamulitrat W.
Zhang X, Gschwind J, Erben V, Bennewitz K, Li X, Sticht C, Poschet G, Hausser I, Fleming T, Szendroedi J, Nawroth PP, Kroll J.
Parstorfer M, Poschet G, Brüning K, Friedmann-Bette B.
Li S, Li H, Bennewitz K, Poschet G, Buettner M, Hausser I, Szendroedi J, Nawroth PP, Kroll J.
Küper K, Poschet G, Rossmann J, Garbade SF, Spiegelhalter A, Wen D, Hoffmann GF, Schmitt CP, Opladen T, Peters V.
Ritterhoff J, McMillen T, Foundas H, Palkovacs R, Poschet G, Caudal A, Liu Y, Most P, Walker M, Tian R.
Zararsiz GE, Lintelmann J, Cecil A, Kirwan J, Poschet G, Gegner HM, Schuchardt S, Guan XL, Saigusa D, Wishart D, Zheng J, Mandal R, Adams K, Thompson JW, Snyder MP, Contrepois K, Chen S, Ashrafi N, Akyol S, Yilmaz A, Graham SF, O'Connell TM, Kalecký K, Bottiglieri T, Limonciel A, Pham HT, Koal T, Adamski J, Kastenmüller G.
Bröker-Lai J, Rego Terol J, Richter C, Mathar I, Wirth A, Kopf S, Moreno-Pérez A, Büttner M, Tan LL, Makke M, Poschet G, Hermann J, Tsvilovskyy V, Haberkorn U, Wartenberg P, Susperreguy S, Berlin M, Ottenheijm R, Philippaert K, Wu M, Wiedemann T, Herzig S, Belkacemi A, Levinson RT, Agarwal N, Camacho Londoño JE, Klebl B, Dinkel K, Zufall F, Nussbaumer P, Boehm U, Hell R, Nawroth P, Birnbaumer L, Leinders-Zufall T, Kuner R, Zorn M, Bruns D, Schwarz Y, Freichel M.
Ott H, Bennewitz K, Zhang X, Prianichnikova M, Sticht C, Poschet G, Kroll J.
Hering M, Madi A, Sandhoff R, Ma S, Wu J, Mieg A, Richter K, Mohr K, Knabe N, Stichling D, Poschet G, Bestvater F, Frank L, Utikal J, Umansky V, Cui G.
Gegner HM, Naake T, Aljakouch K, Dugourd A, Kliewer G, Müller T, Schilling D, Schneider MA, Kunze-Rohrbach N, Grünewald TGP, Hell R, Saez-Rodriguez J, Huber W, Poschet G, Krijgsveld J.
Rades M, Poschet G, Gegner H, Wilke T, Reichert J.
Chausse B, Malorny N, Lewen A, Poschet G, Berndt N, Kann O.
Madai S, Kilic P, Schmidt RM, Bas-Orth C, Korff T, Büttner M, Klinke G, Poschet G, Marti HH, Kunze R.
Ma S, Sandhoff R, Luo X, Shang F, Shi Q, Li Z, Wu J, Ming Y, Schwarz F, Madi A, Weisshaar N, Mieg A, Hering M, Zettl F, Yan X, Mohr K, Ten Bosch N, Li Z, Poschet G, Rodewald HR, Papavasiliou N, Wang X, Gao P, Cui G.
Armbruster L, Pożoga M, Wu Z, Eirich J, Thulasi Devendrakumar K, De La Torre C, Miklánková P, Huber M, Bradic F, Poschet G, Weidenhausen J, Merker S, Ruppert T, Sticht C, Sinning I, Finkemeier I, Li X, Hell R, Wirtz M.
Pfeffer T, Krug SM, Kracke T, Schürfeld R, Colbatzky F, Kirschner P, Medert R, Freichel M, Schumacher D, Bartosova M, Zarogiannis SG, Muckenthaler MU, Altamura S, Pezer S, Volk N, Schwab C, Duensing S, Fleming T, Heidenreich E, Zschocke J, Hell R, Poschet G, Schmitt CP, Peters V.
Riegel G, Orvain C, Recberlik S, Spaety ME, Poschet G, Venkatasamy A, Yamamoto M, Nomura S, Tsukamoto T, Masson M, Gross I, Le Lagadec R, Mellitzer G, Gaiddon C.
Lokumcu T, Iskar M, Schneider M, Helm D, Klinke G, Schlicker L, Bethke F, Müller G, Richter K, Poschet G, Phillips E, Goidts V.
Qian X, Klatt S, Bennewitz K, Wohlfart DP, Lou B, Meng Y, Buettner M, Poschet G, Morgenstern J, Fleming T, Sticht C, Hausser I, Fleming I, Szendroedi J, Nawroth PP, Kroll J.
Mandel N, Büttner M, Poschet G, Kuner R, Agarwal N.
Li H, Seessle J, Staffer S, Tuma-Kellner S, Poschet G, Herrmann T, Chamulitrat W.
Li X, Wu F, Günther S, Looso M, Kuenne C, Zhang T, Wiesnet M, Klatt S, Zukunft S, Fleming I, Poschet G, Wietelmann A, Atzberger A, Potente M, Yuan X, Braun T.
Weisshaar N, Ma S, Ming Y, Madi A, Mieg A, Hering M, Zettl F, Mohr K, Ten Bosch N, Stichling D, Buettner M, Poschet G, Klinke G, Schulz M, Kunze-Rohrbach N, Kerber C, Klein IM, Wu J, Wang X, Cui G.
Li X, Wu F, Günther S, Looso M, Kuenne C, Zhang T, Wiesnet M, Klatt S, Zukunft S, Fleming I, Poschet G, Wietelmann A, Atzberger A, Potente M, Yuan X, Braun T.
Parstorfer M, Poschet G, Kronsteiner D, Brüning K, Friedmann-Bette B.
Pfeffer T, Wetzel C, Kirschner P, Bartosova M, Poth T, Schwab C, Poschet G, Zemva J, Bulkescher R, Damgov I, Thiel C, Garbade SF, Klingbeil K, Peters V, Schmitt CP.
Pfaff DH, Poschet G, Hell R, Szendrödi J, Teleman AA.
Himmelreich N, Kikul F, Zdrazilova L, Honzik T, Hecker A, Poschet G, Lüchtenborg C, Brügger B, Strahl S, Bürger F, Okun JG, Hansikova H, Thiel C.
Dong Y, Silbermann M, Speiser A, Forieri I, Linster E, Poschet G, Samami AA, Wanatabe M, Sticht C, Teleman AA, Deragon JM, Saito K, Hell R, Wirtz M.
Lou B, Wu H, Ott H, Bennewitz K, Wang C, Poschet G, Liu H, Yuan Z, Kroll J, She J.
Qi H, Kan K, Sticht C, Bennewitz K, Li S, Qian X, Poschet G, Kroll J.
Pre-analytical processing of plasma and serum samples for combined proteome and metabolome analysis.
Gegner HM, Naake T, Dugourd A, Müller T, Czernilofsky F, Kliewer G, Jäger E, Helm B, Kunze-Rohrbach N, Klingmüller U, Hopf C, Müller-Tidow C, Dietrich S, Saez-Rodriguez J, Huber W, Hell R, Poschet G, Krijgsveld J.
Tabler CT, Lodd E, Bennewitz K, Middel CS, Erben V, Ott H, Poth T, Fleming T, Morgenstern J, Hausser I, Sticht C, Poschet G, Szendroedi J, Nawroth PP, Kroll J.
Blagojevic B, Almouhanna F, Poschet G, Wölfl S.
Jethva J, Lichtenauer S, Schmidt-Schippers R, Steffen-Heins A, Poschet G, Wirtz M, van Dongen JT, Eirich J, Finkemeier I, Bilger W, Schwarzländer M, Sauter M.
Michalaki A, McGivern AR, Poschet G, Büttner M, Altenburger R, Grintzalis K.
Steimbach RR, Herbst-Gervasoni CJ, Lechner S, Stewart TM, Klinke G, Ridinger J, Géraldy MNE, Tihanyi G, Foley JR, Uhrig U, Kuster B, Poschet G, Casero RA Jr, Médard G, Oehme I, Christianson DW, Gunkel N, Miller AK.
Andresen C, Boch T, Gegner HM, Mechtel N, Narr A, Birgin E, Rasbach E, Rahbari N, Trumpp A, Poschet G, Hübschmann D.
Stewart TM, Foley JR, Holbert CE, Klinke G, Poschet G, Steimbach RR, Miller AK, Casero RA Jr.
Boger M, Bennewitz K, Wohlfart DP, Hausser I, Sticht C, Poschet G, Kroll J.
Gegner HM, Mechtel N, Heidenreich E, Wirth A, Cortizo FG, Bennewitz K, Fleming T, Andresen C, Freichel M, Teleman AA, Kroll J, Hell R, Poschet G.
Dixon LE, Pasquariello M, Badgami R, Levin KA, Poschet G, Ng PQ, Orford S, Chayut N, Adamski NM, Brinton J, Simmonds J, Steuernagel B, Searle IR, Uauy C, Boden SA.
Madi A, Weisshaar N, Buettner M, Poschet G, Ma S, Wu J, Mieg A, Hering M, Ming Y, Mohr K, Ten Bosch N, Cui G.
Alborzinia H, Flórez AF, Kreth S, Brückner LM, Yildiz U, Gartlgruber M, Odoni DI, Poschet G, Garbowicz K, Shao C, Klein C, Meier J, Zeisberger P, Nadler-Holly M, Ziehm M, Paul F, Burhenne J, Bell E, Shaikhkarami M, Würth R, Stainczyk SA, Wecht EM, Kreth J, Büttner M, Ishaque N, Schlesner M, Nicke B, Stresemann C, Llamazares-Prada M, Reiling JH, Fischer M, Amit I, Selbach M, Herrmann C, Wölfl S, Henrich KO, Höfer T, Trumpp A, Westermann F.
Reimer JJ, Shaaban B, Drummen N, Sanjeev Ambady S, Genzel F, Poschet G, Wiese-Klinkenberg A, Usadel B, Wormit A.
Weis EM, Puchalska P, Nelson AB, Taylor J, Moll I, Hasan SS, Dewenter M, Hagenmüller M, Fleming T, Poschet G, Hotz-Wagenblatt A, Backs J, Crawford PA, Fischer A.
Wang J, Fröhlich H, Torres FB, Silva RL, Poschet G, Agarwal A, Rappold GA.





