Multiparametric methods for early detection of prostate cancer
- Imaging and Radiooncology
- Clinical Cooperation Unit
- Junior Research Group

Priv. Doz. Dr. Magdalena Görtz
Group leader
Established in 2021, our Junior-Clinical Cooperation Unit at the DKFZ and the Urology University Hospital Heidelberg is dedicated to addressing highly relevant clinical questions and to drive patient-oriented translational research in prostate cancer. Our primary objectives are personalized early detection and risk stratification, with an emphasis on identifying aggressive prostate cancer, to optimize diagnosis and therapy planning.
Image: © ©Magdalena Görtz/dkfz.de

Image: © ©Magdalena Görtz/dkfz.de
Our Research
Prostate cancer poses a significant challenge due to its varying aggressiveness and progression. This underscores the urgent need for novel molecular markers and integrated diagnostic models that can reliably differentiate aggressive from indolent prostate cancer and benign conditions.
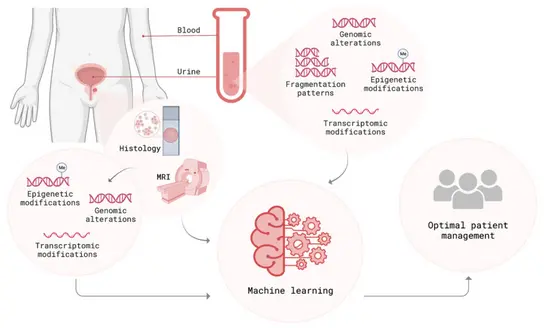
We have prospectively established a comprehensive data cohort and biobank that comprises electronic health records, laboratory results, MRI imaging, digital pathology, (epi)genomic, transcriptomic and proteomic data, as well as patient follow-up information. By integrating multiple data modalities (clinical, laboratory, imaging, as well as liquid- and tissue-derived molecular data) and applying advanced computational techniques, we aim to provide a holistic view of each patient’s disease status, improving early detection and risk stratification for the patient’s benefit. Extracting relevant features from multimodal data and integrating them into a machine learning based prediction model allows for a comprehensive representation of each patient’s tumor at first diagnosis, ultimately personalizing diagnosis and treatment decisions. An integrative early detection and risk stratification strategy that combines clinical parameters, molecular markers and imaging has the potential to optimize non-invasive diagnostics, to develop more precise prediction models and to allow correlations with tumor aggressiveness.
As part of our strong translational approach aimed at clinical impact, we collaborate in consortial projects with academic and industry partners such as the University of Heidelberg and Siemens Healthineers to develop AI-based decision support and “similar-patient search” solutions. These partnerships bridge the gap between clinical practice, academic research, and industry, ensuring that innovative diagnostic and therapeutic approaches have a straight impact on patient care. In line with our patient-centered focus, we developed an AI-based chatbot in partnership with SAP SE, to inform patients about state-of-the-art early detection of prostate cancer. This validated AI-tool exemplifies our direct translation of recent machine learning advances into relevant clinical applications.
Projects
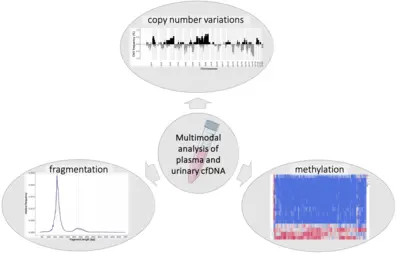
We investigate predictive (epi)genetic modifications in circulating cell-free DNA from prostate cancer patients as a non-invasive method of distinguishing aggressive tumors from indolent prostate cancer and healthy patients.
The multimodal analysis of plasma and urine for non-invasive diagnostics holds great potential for advancing early detection and risk stratification in prostate cancer. Circulating tumor DNA mirrors the heterogeneous molecular status of the primary tumor and its metastasis. Our research group has developed a comprehensive pipeline to evaluate methylation patterns, fragmentation profiles, and copy number alterations in liquid biopsy samples from newly diagnosed prostate cancer patients and cancer-free controls. Multimodal analysis has shown promising results in early cancer detection, as it harnesses synergy between various data types. Our aim is to enable precise molecular characterization and an accurate assessment of tumor aggressiveness at initial diagnosis, supporting optimal, risk-adapted therapeutic decisions for prostate cancer.
Cooperation:
Division of “Cancer Genome Research”, DKFZ
HI-STEM Junior Research Group „Pattern Recognition and Digital Medicine“, DKFZ
Early Cancer Institute, Department of Oncology, University of Cambridge

We combine clinical data, routine blood parameters, and metabolomics to develop non-invasive strategies for the early detection of aggressive prostate cancer.
We prospectively assess whether integrating clinical data with routine blood parameters can improve early, non-invasive prediction of aggressive prostate cancer. Our research identified widely available serum markers that, when combined with traditional PSA testing and clinical factors, significantly increase diagnostic accuracy. The resulting multi-stage risk model outperforms PSA-only methods, offering a cost-effective, non-invasive, and personalized approach for early detection. Using widely available and affordable parameters, our model identified patients at high risk for prostate cancer who require further diagnostic intervention.
In parallel, we are exploring metabolomics as a promising strategy for early detection of aggressive prostate cancer. By comparing metabolomic profiles in blood and urine samples, we aim to identify relevant variations in metabolite concentrations among patients with aggressive prostate cancer, indolent disease, and healthy individuals.
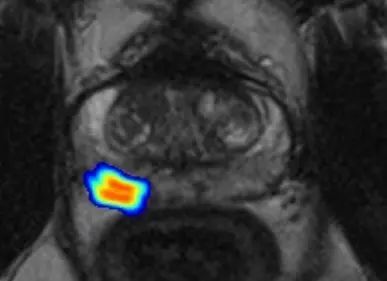
By extracting quantitative features from MRI data, integrating clinical and molecular data and applying machine learning technologies, we uncover correlations between imaging findings and underlying tumor biology.
Given the limited sensitivity and specificity of current early detection methods for prostate cancer, integrating multiple data sources is crucial for achieving more accurate diagnostics. Our projects focus on combining clinical data and advanced imaging techniques to optimize decision-making in prostate cancer care. This includes the integration of clinical and imaging data for personalized early detection and patient follow-up strategies, as well as improving MRI-ultrasound fusion during prostate biopsies for precision diagnostics.
To further refine non-invasive screening, our research aims to improve risk assessment and reduce unnecessary prostate biopsies by applying deep learning to imaging data and integrating these findings with clinical and molecular data. By merging insights from clinical and imaging findings with molecular data from liquid biopsies, we aim to establish more precise, non-invasive diagnostic strategies. Radiomic features extracted from MRI scans serve as inputs for machine-learning algorithms and are correlated with fragmentation profiles and markers of genomic instability obtained from liquid biopsy samples. This comprehensive integration of clinical data, radiomic features, and molecular markers not only enhances non-invasive assessment of prostate cancer but also provides deeper insights into genotype-phenotype correlations, advancing the field of precision diagnostics.
Cooperation:
Division of “Radiology”, DKFZ

Using spatial proteomics and transcriptomics, we characterize the tumor microenvironment at high-resolution, identifying markers of aggressive prostate cancer at first diagnosis.
This research project focuses on early-stage prostate cancer, applying spatial transcriptomics and proteomics techniques, including cutting-edge single-cell spatial imaging platforms, to provide an unparalleled, spatially resolved view of prostate tumor cell heterogeneity and their intratumoral microenvironment.
To analyze these spatial datasets, we employ advanced bioinformatics techniques such as machine learning algorithms, multi-omics data integration, and high-dimensional data visualization. These methods help us identifiy critical molecular interactions and pathways that drive disease progression. We aim to integrate these findings into clinically relevant stratification models, enabling precise risk assessments and informing personalized treatment recommendations for patients with early-stage prostate cancer. The research group’s prospectively established study cohort, including comprehensive clinical data, oncological follow-up, and imaging, ensures robust integration and translation of both molecular and clinical insights.
Cooperation:
Division of “Computational Genomics and Systems Genetics”, DKFZ
Department of Pathology, Heidelberg University Medical Center
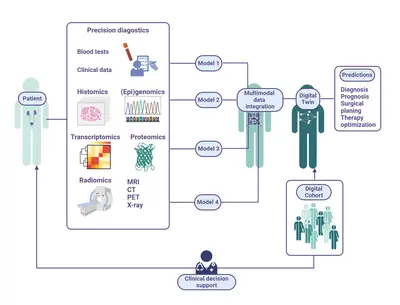
We integrate molecular, imaging, and clinical data into a “digital twin” of prostate cancer patients, leveraging AI to capture patient-specific, multidimensional health data and reflect real-world tumor characteristics.
For many years, we have been cooperating with academic and industry partners, focusing on precision therapy and clinical translation. This collaboration enabled us to secure sustainable funding from the German Federal Ministry for Economic Affairs and Climate Action, providing a strong foundation for our translational prostate cancer research. As part of these efforts, the Junior Clinical Cooperation Unit played an integral part in the CLINIC 5.1 consortium (Home (clinic51.de)). This initiative brought together the German Cancer Research Center, Heidelberg University Hospital, Heidelberg University, KARL STORZ, SAP SE, and Siemens Healthineers AG to develop innovative, market-oriented AI-based decision support tools for physicians. The consortium‘s focus was to improve decision-making and precision medicine in prostate cancer diagnosis, treatment planning, and therapy. By integrating and expanding diagnostic and therapeutic datasets, the CLINIC 5.1 project aimed to develop a comprehensive, four-dimensional virtual representations of prostate cancer patients („digital twins“).
Recently, a collaboration with the Engineering Mathematics and Computing Lab (Interdisciplinary Center for Scientific Computing, Heidelberg University) was launched to advance multimodal analysis of prostate cancer. The project centers on three key objectives: employing partial differential equations to model tumor growth, developing a multimodal AI framework to estimate tumor recurrence probabilities with uncertainty-aware regression models, and ensuring the interpretability of multimodal AI models to build clinician trust. An integral mission of this collaboration is the strive for precision oncology through innovative, multidisciplinary approaches with a robust mathematical foundation.
Team
A dedicated, cross-disciplinary team of experts in medicine, molecular biology, biotechnology, and bioinformatics works in synergy to advance non-invasive early detection of prostate cancer and refine risk stratification at initial diagnosis, ensuring optimal treatment decisions for patients.
- Show profile

Priv. Doz. Dr. Magdalena Görtz
Group leader
-

Dr. Anja Lisa Riediger
PhD Studentin
-

Dr. Martina Heller
Clinician Scientist
-

Isabella Schindler
Wissenschaftliche Mitarbeiterin M.Sc.
-

Daniela Janscho
Study Nurse
Optimizing early prostate cancer detection: the research team’s goals and personalized approach
In this video, PD Dr. Magdalena Görtz explains the importance of an individualized approach in the early detection of prostate cancer and presents innovative methods that her research team is developing to optimize patient treatment.

Funding
We gratefully acknowledge the financial support from the following funding bodies for helping us to carry out our research projects:

Selected publications
Eickelschulte S, Kaczorowski A, Janke F, Riediger AL, Lazareva O, Böning S, Kristiansen G, Schwab C, Stenzinger A, Sültmann H, Duensing S, Duensing A, Görtz M
Lazareva O, Riediger A, Stegle O, Sültmann H, Hohenfellner M, Görtz M
Baumgärtner K, Byczkowski M, Schmid T, Muschko M, Woessner P, Gerlach A, Bonekamp D, Schlemmer HP, Hohenfellner M, Görtz M
Janke F, Stritzke F, Dvornikovich K, Franke H, Angeles AK, Riediger AL, Ogrodnik S, Gerhardt S, Regnery S, Schröter P, Bauer L, Weusthof K, Görtz M, Harrabi S, Herfarth K, Neelsen C, Paech D, Schlemmer HP, Abdollahi A, Adeberg S, Debus J, Sültmann H, Held T.
Schrader A, Netzer N, Hielscher T, Görtz M, Zhang KS, Schütz V, Stenzinger A, Hohenfellner M, Schlemmer HP, Bonekamp D
Lleshi E, Milne-Clark T, Lee Yu H, Martin HW, Hanson R, Lach R, Rossi SH, Riediger AL, Görtz M, Sültmann H, Flewitt A, Lynch AG, Gnanapragasam VJ, Massie CE
Görtz M, Baumgärtner K, Schmid T, Muschko M, Woessner P, Gerlach A, Byczkowski M, Sültmann H, Duensing S, Hohenfellner M
Eickelschulte S, Riediger AL, Angeles AK, Janke F, Duensing S, Sültmann H, Görtz M
Tavakoli AA, Hielscher T, Badura P, Görtz M, Kuder TA, Gnirs R, Schwab C, Hohenfellner M, Schlemmer HP, Bonekamp D
Görtz M, Hohenfellner M
Görtz M, Radtke JP, Hatiboglu G, Schutz V, Tosev G, Guttlein M, Leichsenring J, Stenzinger A, Bonekamp D, Schlemmer HP
Further publications
Get in touch with us
