Mechanisms of Genomic Variation and Data Science
- Functional and Structural Genomics

Prof. Dr. Jan Korbel
The vision of our research program is to understand genomic structural variants (SVs) such as copy-number variants, translocations and patterns of complex DNA rearrangement as a basis of human phenotypic variation and cancer.

Our Research
SVs are responsible for most varying nucleotide bases between individual human genomes. SVs also emerge somatically in most cancers, where they comprise the primary form of driver mutation outnumbering point mutation drivers, result in vast genetic heterogeneity, contribute to tumorigenesis, and affect therapy response. Our research interest is to uncover determinants behind the formation and selection of SVs.
Through my active involvement in data science activities, I am also facilitating the bridging of biological and clinical research, via data sharing and application of cutting - edge methods in data science, artificial intelligence and machine learning (AI/ML). Further to his role at the DKFZ, Jan Korbel is primary affiliated with the European Molecular Biology Laboratory (EMBL) in the Genome Biology Unit, and is contributing to a strategic bridge in data science between both institutions relevant to genomic data sharing and data science frameworks.
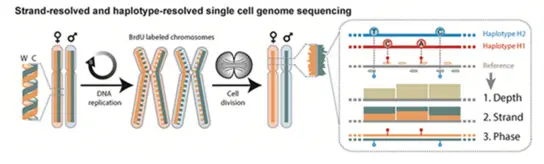
Our group will develop methods to integrate high-resolution bioimaging and single cell sequencing (Strand-seq) using cutting edge methods in data science and machine learning, in order to dissect SV formation mechanisms, and to facilitate a long needed convergence of cell biology and genomics through integrating confocal microscopy and single cell multi-omics approaches.
Team
-

Prof. Dr. Jan Korbel
-
Eva Benito
-
Carsten Hain
-

Dr. Maise Queiroz
-
Tobias Rausch
-

Catherine Stober
Selected publications
Recurrent inversion polymorphisms in humans associate with genetic instability and genomic disorders
Porubsky, D., Höps, W., Ashraf, H., Hsieh, P., Rodriguez-Martin, B., Yilmaz, F., Ebler, J., Hallast, P., Maria Maggiolini, F.A., Harvey, W.T., Henningm, B., Audano, P.A., Gordon, D.S., Ebert, P., Hasenfeld, P., Benito, E., Zhu, Q.; Human Genome Structural Variation Consortium (HGSVC), Lee, C., Antonacci, F., Steinrücken, M., Beck, C.R., Sanders, A.D., Marschall, T., Eichler, E.E., Korbel, J.O.
Jeong, H., Grimes, K., Bruch, P., Rausch, T., Hasenfeld, P., Sabarinathan, R., Porubsky, D., Herbst, S.A., Erarslan-Uysal, B., Jann, J., Marschall, T., Nowak, D., Bourquin, J., Kulozik, A.E., Dietrich, D., Bornhauser, B., Sanders, A.D., Korbel, J.O.
Single cell tri-channel-processing reveals structural variation landscapes and complex rearrangement processes
Sanders, A.D., Meiers, S., Ghareghani, M., Porubsky, D., Jeong, H., van Vliet, A.M.C.C., Rausch, T., Richter-Pechanska, P., Kunz, J.B., Jenni, S., Bolognini, D., Longo, G.M.C., Raeder, B., Kinanen, V., Zimmermann, J., Benes, V., Schrappe, M., Mardin, B.R., Kulozik, A., Bornhauser, B., Bourquin, J., Marschall T, Korbel, J.O.
ICGC/TCGA Pan-Cancer Analysis of Whole Genomes Consortium (PCAWG)
Get in touch with us

