Research
- Research Topics
- Cell Biology and Tumor Biology
- Stem Cells and Cancer
- Inflammatory Stress in Stem Cells
- Experimental Hematology
- Molecular Embryology
- Signal Transduction and Growth Control
- Epigenetics
- Redox Regulation
- Vascular Oncology and Metastasis
- Clinical Neurobiology
- Molecular Neurogenetics
- Chaperones and Proteases
- Vascular Signaling and Cancer
- Molecular Neurobiology
- Mechanisms Regulating Gene Expression
- Molecular Biology of Centrosomes and Cilia
- Dermato-Oncology
- Pediatric Leukemia
- Tumour Metabolism and Microenvironment
- Personalized Medical Oncology
- Molecular Hematology - Oncology
- Cancer Progression and Metastasis
- Translational Surgical Oncology
- Neuronal Signaling and Morphogenesis
- Cell Signaling and Metabolism
- Cell Fate Engineering and Disease Modeling
- Cancer Drug Development
- Cell Morphogenesis and Signal Transduction
- Functional and Structural Genomics
- Molecular Genome Analysis
- Molecular Genetics
- Pediatric Neurooncology
- Cancer Genome Research
- Chromatin Networks
- Functional Genome Analysis
- Theoretical Systems Biology
- Neuroblastoma Genomics
- Signaling and Functional Genomics
- Signal Transduction in Cancer and Metabolism
- RNA Biology and Cancer
- Systems Biology of Signal Transduction
- Areas of Interest
- Advancement of clinical proteomics for systems medicine
- Bridging from the single cell to the cell population – Epo-induced cellular responses and erythroleukemia
- Deciphering tumor microenvironment interactions determining lung cancer development
- Mechanisms controlling the compensation of liver injury and towards model-based biomarkers for early detection of liver cancer
- Application of dynamic pathway modelling for personalized medicine
- Group Members
- Publications
- Open Positions
- Funding
- Teaching
- Areas of Interest
- Molecular thoracic Oncology
- Proteomics of Stem Cells and Cancer
- Computational Genomics and System Genetics
- Applied Functional Genomics
- Applied Bioinformatics
- Translational Medical Oncology
- Metabolic crosstalk in cancer
- Pediatric Glioma Research
- Cancer Epigenomics
- Translational Pediatric Sarcoma Research
- Artificial Intelligence in Oncology
- Mechanisms of Genomic Variation and Data Science
- Neuropathology
- Pediatric Oncology
- Neurooncology
- Somatic Evolution and Early Detection
- Translational Control and Metabolism
- Soft-Tissue Sarcoma
- Precision Sarcoma Research
- Brain Mosaicism and Tumorigenesis
- Mechanisms of Genome Control
- Translational Gastrointestinal Oncology and Preclinical Models
- Translational Lymphoma Research
- Mechanisms of Leukemogenesis
- Genome Instability in Tumors
- Developmental Origins of Pediatric Cancer
- Brain Tumor Translational Targets
- Translational Functional Cancer Genomics
- Regulatory Genomics and Cancer Evolution
- SPRINT
- Cancer Risk Factors and Prevention
- Cancer Epidemiology
- Biostatistics
- Clinical Epidemiology and Aging Research
- Health Economics
- Physical Activity, Prevention and Cancer
- Preventive Oncology
- Personalized Early Detection of Prostate Cancer
- Digital Biomarkers for Oncology
- Genomic Epidemiology
- Cancer Survivorship
- Immunology and Cancer
- Cellular Immunology
- Molecular Oncology of Gastrointestinal Tumors
- Immunoproteomics
- T Cell Metabolism
- Personalized Immunotherapy
- mRNA Cancer Immunotherapies
- Translational Immunotherapy
- B Cell Immunology
- Immune Diversity
- Structural Biology of Infection and Immunity
- Applied Tumor Immunity
- Neuroimmunology and Brain Tumor Immunology
- Adaptive Immunity and Lymphoma
- Immune Regulation in Cancer
- Systems Immunology and Single Cell Biology
- GMP & T Cell Therapy
- Immune Monitoring
- News
- Imaging and Radiooncology
- Radiology
- Research
- Computational Radiology Research Group
- Contrast Agents In Radiology Research Group
- Neuro-Oncologic Imaging Research Group
- Radiological Early Response Assessment Of Modern Cancer Therapies
- Imaging In Monoclonal Plasma Cell Disorders
- 7 Tesla MRI - Novel Imaging Biomarkers
- Functional Imaging
- Visualization And Forensic Imaging
- PET/MRI
- Dual- and Multienergy CT
- Radiomics Research Group
- Prostate Research Group
- Breast Imaging Research Group
- Bone marrow
- Musculoskeletal Imaging
- Microstructural Imaging Research Group
- Staff
- Patients
- Research
- Medical Physics in Radiology
- X-Ray Imaging and Computed Tomography
- Federated Information Systems
- Translational Molecular Imaging
- Medical Physics in Radiation Oncology
- Biomedical Physics in Radiation Oncology
- Intelligent Medical Systems
- Medical Image Computing
- Radiooncology - Radiobiology
- Smart Technologies for Tumor Therapy
- Radiation Oncology
- Molecular Radiooncology
- Nuclear Medicine
- Translational Radiation Oncology
- Molecular Biology of Systemic Radiotherapy
- Interactive Machine Learning
- Multiparametric methods for early detection of prostate cancer
- Molecular Mechanisms of Head and Neck Tumors
- Radiology
- Infection, Inflammation and Cancer
- Tumor Virology
- Viral Transformation Mechanisms
- Pathogenesis of Virus-Associated Tumors
- Immunotherapy and Immunoprevention
- Applied Tumor Biology
- Virotherapy
- Virus-associated Carcinogenesis
- Chronic Inflammation and Cancer
- Microbiome and Cancer
- Cell Plasticity and Epigenetic Remodeling
- Experimental Hepatology, Inflammation and Cancer
- Infections and Cancer Epidemiology
- Tumorvirus-specific Vaccination Strategies
- Mammalian Cell Cycle Control Mechanisms
- Molecular Therapy of Virus-Associated Cancers
- DNA Vectors
- Episomal-Persistent DNA in Cancer- and Chronic Diseases
- Cell Biology and Tumor Biology
- Research Groups A-Z
- Junior Research Groups
- Core Facilities
- Center for Preclinical Research
- Chemical Biology Core Facility
- Electron Microscopy
- Flow Cytometry
- Genomics and Proteomics
- Information Technology
- Library
- Kataloge -- Catalogues
- Zeitschriften - Journals
- E-Books - ebooks
- Datenbanken - Databases
- Dokument-Lieferung - Document Delivery
- Publikationsdatenbank - Publication database
- DKFZ Archiv - DKFZ Archive
- Open Access
- Science 2.0
- Ansprechpartner - Contact
- More Information - Service
- Anschrift - Address
- Antiquariat - Second Hand
- Aufstellungssystematik - Shelf Classification
- Ausleihe - Circulation
- Benutzerhinweise - Library Use
- Beschaffungsvorschläge - Desiderata
- Fakten und Zahlen - Facts and Numbers
- Kooperationen, Konsortien - Cooperations, Consortia
- Kopieren, Scannen - Copying, Scans
- Kurse, Führungen - Courses, Introductions
- DKFZ-Intern - internal only
- DEAL-Info
- Light Microscopy
- Omics IT and Data Management Core Facility
- Small Animal Imaging
- Metabolomics Core Technology Platform
- Data Science @ DKFZ
- INFORM
- Baden-Württemberg Cancer Registry
- Cooperations & Networks
- National Cooperations
- International Cooperations
- Cooperational Research Program with Israel: DKFZ - MOST in Cancer Research
- Program
- Members of the Program Committee
- Call
- Publication Database
- German-Israeli Cancer Research Schools
- Archive
- Heidelberg - Israel, Science and Culture
- Symposium 40 Years of German-Israeli Cooperation
- 35th Anniversary Symposium
- 34th Meeting of the DKFZ-MOST Program
- 40th Anniversary Publication
- 30th Anniversary Publication
- 20th Anniversary Publication
- Flyer - The Cancer Cooperation Program
- List Publications 1976-2004
- Highlight-Projects
- Cooperational Research Program with Israel: DKFZ - MOST in Cancer Research
- Cooperations with industrial companies
- DKFZ PostDoc Network
- Cross Program Topic RNA@DKFZ
- Cross Program Topic Epigenetics@dkfz
- Cross Program Topic Single Cell Sequencing
- WHO Collaborating Centers
- DKFZ Site Dresden
- Health + Life Science Alliance Heidelberg Mannheim
Oncogenomics
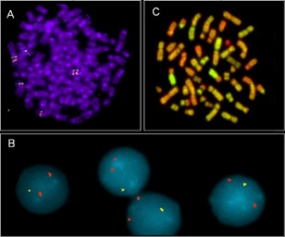
Fig.1
© dkfz.de
In order to identify the critical genes which are involved in the development of malignant cells we analyze chromosomal aberrations in clinically well defined tumor samples by means of fluorescence in situ hybridization (FISH) (Fig. 1 A, B). Furthermore, we use Comparative Genomic Hybridization (CGH) as a genomic screening approach in order to identify recurrent numerical chromosomal changes, by which oncogenes or tumorsupressor genes could be activated (1c). The results of our analyses are complemented in our department by further approaches, like Matrix-CGH, expression profiling, functional tumor genetics and bioinformatic analysis.
Fig.1: Analysis of chromosomal changes in tumor cell genomes by fluorescence in-situ hybridization (FISH). (A) FISH on a metaphase spread of Hodgkin lymphoma cells. The example shows the duplication of a region of chromosome 2 carrying the c-Rel oncogene. (B) FISH analysis on interphase nuclei (interphase cytogenetics). An experiment on interphase nuclei from chronic lymphocytic leucaemia (CLL) cells is shown. Two different DNA probes, one from a locus on chromosome 7 (red) and one from the locus of tumorsuppressor gene RB-1 (green) were hybridized. While the chromosome 7 derived probe does not show a numerical change (i.e. 2 copies per cell) the RB-1 probe shows only 1 signal, i.e. the RB-1 locus is deleted in these cells. (C) Example of a CGH experiment. Total genomic DNA of tumor cells (green) together with a differently labelled genomic control DNA from normal cells (red) is hybridized on normal metaphase chromosomes. Chromosomal gains or losses are assessed by the hybridization intensity ratio between tumor and normal DNA. Chromosomal gains within the tumor cells result in an increased intensity ratio between tumor and control DNA (greenish staining), losses in a decreased ratio between tumor and control DNA (reddish staining) of the corresponding target chromosomes.
selected publications
- Mapping and chromosome analysis: the potential of fluorescence in situ hybridization.
- Detection of chromosomal aberrations by means of molecular cytogenetics: painting of chromosomes and chromosomal subregions and comparative genomic hybridization.
- Comparative genomic hybridization: uses and limitations.
- New tools in molecular pathology.
