New method can detect gene therapy vectors in clinical trials
Modeling the genomic accessibility of viral integration sites
© dkfz.de
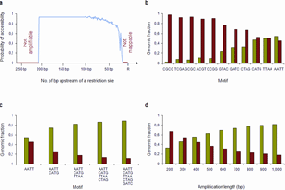
Once a new drug has been developed, to understand how it works in human patients requires to determine the distribution of the medication present in the body. This is often achieved by measuring the level of the drug and its byproduct in the blood and urine over the course of time. Such kinetics of pharmaceutical drugs in an organism, termed pharmacokinetics, have been very difficult or impossible to determine in gene therapies, even though it would be especially important to understand the whereabouts of gene vectors in this new treatment modality. In a novel study published in this month’s edition of Nature Medicine, Gabriel and colleagues from the National Center for Tumor Diseases (NCT) at the German Cancer Research Center in Heidelberg, Germany have modeled how gene therapy vectors distribute after therapeutic applications in the genome of the treated cells, and describe a method to retrieve vectors and mutations in the entire human genome.
Retroviral vectors have been used very successfully to treat immunodeficiencies, but have also induced side effects. Some degree of subtle influence on the growth of white blood cells can be detected in many gene therapy patients. Severe clonal proliferation and leukemia have required intensive further treatment in some of them. These observations emphasize the need for comprehensive analyses to assess the pharmacokinetics of vectors in the cells of patients. Gabriel and colleagues from the group of Christof von Kalle have generated a snapshot of the human genome that shows which areas escape analysis for the presence of therapeutic gene vectors. Their work shows that previous methods to address this issue use enzymatic restriction that has often only allowed to find a fraction of all genomic integrants of gene therapy vectors, hampering the understanding and prediction of biological consequences after vector insertion. The new approach described in the Paper in Nature medicine has superior capabilities for comprehensively detecting where vector integrations can be located in the entire genome of cells. It is a decisive step forward in understanding and thereby improving the use of gene vectors to treat genetic diseases, including inborn defects of the immune system, cancer and HIV infection.
Gabriel, R., Eckenberg, R., Paruzynski, A., Bartholomae, C., Nowrouzi, A., Arens, A., Howe, S.J., Recchia, A., Cattoglio, C., Wang, W., Faber, K., Schwarzwaelder, K., Kirsten, R., Deichmann, A., Ball, C.R., Balaggan, K.S., Yáñez-Muñoz, R.J., Ali, R.R., Gaspar, H.B., Biasco, L., Aiuti, A., Cesana, D., Montini, E., Naldini, L., Cohen-Haguenauer, O., Mavilio, F., Thrasher, A.J., Glimm, H., von Kalle, C., Saurin, W., Schmidt, M. Comprehensive genomic access to vector integration in clinical gene therapy. Nature Medicine. 2009 Dec;15(12):1431-6. Epub 2009 Nov 22. DOI:10.1038/nm.2057
