Regulatory Genomics and Cancer Evolution
- Functional and Structural Genomics

Prof. Dr. Duncan Odom
Group Leader
Dr Odom’s laboratory studies how genetic sequence information shapes the cell's DNA regulatory landscape and thus the trajectory of cancer genome evolution.
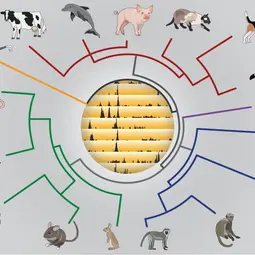
Our Research
We are mainly an experimental genomics laboratory with long-standing collaborative alliances with other - particularly computational - research groups. The breadth of our three major scientific themes provides substantial latitude to tailor particular projects to the scientific interest of incoming students and postdocs.
First, we are testing how genetic sequence variation shapes genome regulation and gene expression in phenotypically normal somatic tissues. One strategy is to profile and compare regulatory architectures and function among closely- and distantly-related mammals. Our laboratory is known for its early pioneering application of interspecies comparisons to reveal the extensive and rapid turn-over of tissue-specific transcription factor binding, CTCF/insulator elements, polymerase occupancies, and enhancer activities. We are actively developing novel approaches to integrate recently developed single-cell transcriptional, spatial, and epigenomic datasets [funded by an ERC Advanced Grant].
Second, we are establishing the earliest mechanisms of cancer genome formation. We are use chemical carcinogenesis to create and analyse liver tumours in multiple different mammalian species. This approach is the first artificially created and carefully controlled tumour cohorts that can be reliably and quantitatively compared, in order to reveal the underlying principles of cancer genome evolution. Our work has recently discovered that chemical carcinogen-driven tumours can carry megabase scale mutational asymmetries, and we are exploiting this to test how mutations are first established in the cancer genome.
Third, we are exploring how ageing interacts with genetic diversity in shaping genome stability. We exploit closely related mouse species with a similar organismal phenotype, but with highly divergent genomes. We have used this strategy to begin exploring how tissue function changes in the bone marrow and the female reproductive tract. Recent published studies from the Odom lab have used single-cell transcriptional analysis to demonstrate how ageing can result in substantial increases in cell-to-cell transcriptional variability in the immune system.
Our Team
- Show profile

Prof. Dr. Duncan Odom
Group Leader
- Show profile

Dr. Paul Ginno
Staff Scientist
- Show profile

Dr. Stefania Del Prete
PostDoc
- Show profile

Dr. James Cleland
PostDoc
- Show profile

Klaudija Daugelaite
PostDoc
- Show profile

Dr. Veronica Busa
PostDoc
- Show profile
Dr. Jusung Lee
PostDoc
- Show profile

Dr. Raphael Teodoro Franciscani Coimbra
Bioinformatician
- Show profile

Anja Schneider
Lab Manager
- Show profile

Marie-Luise Koch
Lab Manager
- Show profile
Nina Wilhelm
Lab Technician
- Show profile

Lilla Pecori
Secretary
- Show profile
Eileen Haage
Secretary
- Show profile

Milica Bekavac
PhD Student
- Show profile

Eleonore Boiral
PhD Student
- Show profile
Michele Framarin
PhD Student
- Show profile

Irem Tellioglu Akcay
PhD Student
- Show profile

Lea Christine Wölbert
PhD Student
Friends from other Labs
- Show profile

Dr. Ivana Winkler
PostDoc
- Show profile

Perrine Lacour
PhD Student
- Show profile

Anna Mathioudaki
PostDoc
- Show profile

Domenico Calafato
PhD Student
- Show profile

Heng Luo
PhD Student
-
Selected Publications
Milica Bekavac, Raphael Coimbra, Veronica F Busa, Mikaela Behm, Rebecca E Wagner, Angela Goncalves, Sabine Begall, Michaela Frye, Duncan T Odom
Klaudija Daugelaite, Perrine Lacour, Ivana Winkler, Marie-Luise Koch, Anja Schneider, Nina Schneider, Francesca Coraggio, Alexander Tolkachov, Xuan Phuoc Nguyen, Adriana Vilkaite, Julia Rehnitz, Duncan T Odom, Angela Goncalves
Panten, J.* ; Del Prete, S. *; Cleland, J. ; Saunders, L. M.* ; van Riet, J.* ; Schneider, A.* ; Ginno, P.* ; Schneider, N.* ; Koch, M.-L.* ; Chen, X. ; Gerstung, M.* ; Stegle, O.* ; Arnold, A. P. ; Turner, J. M. A. ; Heard, E. ; Odom, D.*
Winkler, I.* ; Tolkachov, A.* ; Lammers, F.* ; Lacour, P.* ; Daugelaite, K.* ; Schneider, N.* ; Koch, M.-L.* ; Panten, J.* ; Grünschläger, F.* ; Poth, T. ; Ávila, B. M. d. ; Schneider, A. ; Haas, S.* ; Odom, D. T.* ; Gonçalves, Â.*
Anderson, C. J. ; Talmane, L. ; Ginno, P. A.*; Semple, C. A. ; Odom, D.* ; Aitken, S. J. ; Taylor, M. S.; et al.
Ginno, P. ; Borgers, H.* ; Ernst, C. ; Schneider, A.* ; Behm, M.* ; Aitken, S. J. ; Taylor, M. S. ; Odom, D. *
Get in touch with us

Prof. Dr. Duncan Odom
Group LeaderPostal address:



