Translational Neuroblastoma Research
- Functional and Structural Genomics
- KiTZ

Priv. Doz. Dr. Frank Westermann
Group Leader
Overall aim of our work is to integrate next-generation molecular diagnostics for a more precise patient risk stratification and to develop molecularly targeted therapies based on a better understanding of the molecular mechanisms underlying neuroblastoma tumorigenesis.
Image: Telomere FISH of CHLA-90 neuroblastoma cells,
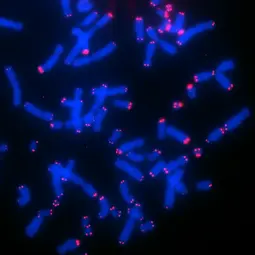
Image: Telomere FISH of CHLA-90 neuroblastoma cells,
Our Research
Neuroblastoma (NB) is the most common pediatric single-entity solid cancer derived from primitive cells of the peripheral sympathetic nervous system. It is characterized by heterogeneous clinical phenotypes ranging from spontaneous regression to malignant progression despite intensive multimodal therapies. The presence of an active telomere maintenance mechanism is associated with aggressive growth and poor outcome in neuroblastoma, while low-risk tumors usually lack a telomere maintenance mechanism. Subsets of high-risk neuroblastomas elongate telomeres either by telomerase activation (as a result of amplified MYCN or rearranged TERT) or by alternative lengthening of telomeres (ALT).
We use multi-omics high-throughput profiling with the specific aim to define genetic, epigenetic and metabolic alterations associated with distinct neuroblastoma subtypes. Furthermore, we aim at understanding the enhancer regulated core regulatory circuitries defining the epigenome and transcriptome of neuroblastoma cells and their role in oncogenesis, relapse and metastasis formation. With the help of single-cell sequencing technologies in combination with spatial transcriptomics of tumor cells in comparison to normal embryonal and fetal tissues during development, we investigate the developmental origins and evolutionary trajectories of neuroblastoma tumors. Our (epi)genetic work is complemented by the study of deregulated metabolic networks in neuroblastoma cells and how these can provide an Achilles heel for cancer therapy.
To disclose neuroblastoma-specific vulnerabilities associated with distinct genetic aberrations and to systematically identify specific targets for personalized medicine approaches, we use high-throughput genetic and compound screens. We have a long-standing expertise in preclinical target development and validation in close collaboration with clinical and industry partners.
Projects
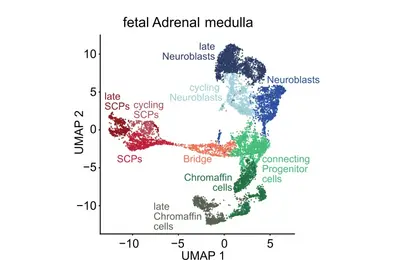
Genomic and other molecular analyses in different cancer types have revealed a striking diversity of genomic aberrations, altered signaling pathways and oncogenic processes. We hypothesize that this diversity arises from endogenous factors, including developmental programs and epigenetic states of the originating cells, in conjunction with exogenous factors. A precise definition of the cell-of-origin and differentiation/developmental states during key events is of utmost importance. The aim of this research focus is to identify resurrected developmental programs in individual tumor cells by comparison to normal human embryonic/fetal cells.
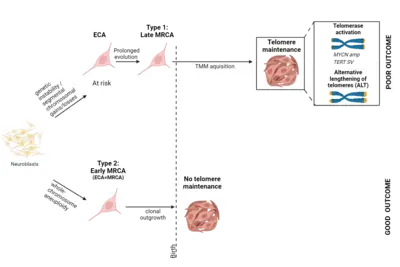
A central question of our work is how specific mutations alter the normal development of the tissue of origin. The goal of this research focus is to understand exactly when neuroblastomas begin to form during embryonic development and how benign tumors differ from highly aggressive ones. Our analyses suggest that neuroblastomas likely begin to form as early as the first trimester of pregnancy, across the entire clinical spectrum. Using specific genetic alterations and mathematical models, we are able to reconstruct the developmental history of each individual tumor. This calculation is based on the assumption that genetic alterations accumulate randomly in the genome over time at a constant rate—similar to sand in an hourglass. The accumulation of mutations is thus also referred to as a molecular clock, which is measurable and allows us to draw conclusions about the temporal history of tumor development. Surprisingly, the analyses revealed that neuroblastomas from all risk groups arise in early pregnancy, but differ in the nature and duration of their early genetic evolution (Körber et al., Nature Genetics 2022).

To better understand the onset and development of neuroblastoma, we model key (epi)genetic events, that we have identified in the tumors, in human induced pluripotent stem cells (iPSCs). These iPSCs can be differentiated in the laboratory into various developmental stages of embryonic development. This allows us to test in which cell populations specific (epi)genetic alterations contribute to the formation of neuroblastoma.
Ferroptosis is a form of cell death triggered when a lethal amount of free radicals accumulates within the cell. This occurs when the metabolic systems that regulate the balance of oxygen, iron, amino acids, and polyunsaturated fatty acids fail. Neuroblastoma cells are particularly vulnerable to ferroptosis due to their high metabolic activity. However, cancer cells have developed specific mechanisms to protect themselves from cell death by ferroptosis. The goal of our work is to understand the complex regulatory system in neuroblastoma cells and, by doing so, identify potential targets to selectively trigger ferroptosis and develop new therapeutic concepts. It is also crucial to understand how the regulatory systems differ across the various molecular subtypes of neuroblastoma, leading to different sensitivities to these therapies. Our research on ferroptosis is funded by the German Research Foundation (DFG) as part of the priority program SPP 2306, titled "Ferroptosis: From Molecular Foundations to Clinical Applications."
An important goal of our work for many years has been to identify target molecules in neuroblastoma cells that depend on high activity of the MYC(N) oncogene, since MYC(N) itself cannot easily be therapeutically targeted. Based on a siRNA screen, we identified the inhibition of the CDK12/13/CCNK complex as a potential target. In a drug screening, we identified various chemical substances that specifically target the CDK12/13/CCNK complex. These substances are based on a very similar chemical structure but have different modes of action. Some substances act as classical kinase inhibitors, while others function as so-called molecular glue degraders. These molecules exploit the cell's ubiquitin-proteasome system to selectively degrade their target proteins, thereby rendering them ineffective. Our work is supported within the framework of the international PROTECT project, which is part of the "Cancer Grand Challenges" program. The developed substances are clinically very promising and are being thoroughly tested preclinically for efficacy and safety.
Our group is part of the international project ITCC pediatric cancer data portal, which is an international partnership involving clinical programs spanning seven nations aiming to aggregate and harmonize genomic data from over 6000 pediatric cancer cases.
For more information see the project website: https://www.pedcanportal.eu/
Team
Our team is composed of wet lab and bioinformatic scientists as well as technical staff.
-

Priv. Doz. Dr. Frank Westermann
Group Leader
-
Dr. Kai-Oliver Henrich
Senior Scientist
-
Dr. Sina Kreth
Senior Scientist
-

Dr. Anand Mayakonda Thippeswamy
Group Leader Bioinformatics
-
Robin Droit
Postdoc
-

Dr. Sabine Stainczyk
Project Manager
-

Cedar Schloo
PhD Student
-
Ilayda Özel
PhD Student
-
Pravin Velmurugan
PhD Student
-

Michael Müller
MD/PhD Student
-
Pascal Kohmann
MD Student
-
Yeo-Eun Yi
MD Student
-
Maximilia Eggle
MD Student
-

Young-Gyu Park
Lab Manager
-

Elisa Maria Wecht
Lab Technician - BSc
-
Michelle Müller
Trainee Lab Technician
-
Adrianna Podolak
Master Student
-
Manas Sehgal
Master Student
Selected Publications
Körber, V., S. A. Stainczyk, R. Kurilov, K. O. Henrich, B. Hero, B. Brors, F. Westermann* and T. Höfer*
Alborzinia, H., A. F. Florez, S. Kreth, L. M. Bruckner, U. Yildiz, M. Gartlgruber, D. I. Odoni, G. Poschet, K. Garbowicz, C. Shao, C. Klein, J. Meier, P. Zeisberger, M. Nadler-Holly, M. Ziehm, F. Paul, J. Burhenne, E. Bell, M. Shaikhkarami, R. Wurth, S. A. Stainczyk, E. M. Wecht, J. Kreth, M. Buttner, N. Ishaque, M. Schlesner, B. Nicke, C. Stresemann, M. Llamazares-Prada, J. H. Reiling, M. Fischer, I. Amit, M. Selbach, C. Herrmann, S. Wolfl, K. O. Henrich, T. Höfer*, A. Trumpp*, and F. Westermann*
Single-cell transcriptomic analyses provide insights into the developmental origins of neuroblastoma
Jansky, S., A. K. Sharma, V. Körber, A. Quintero, U. H. Toprak, E. M. Wecht, M. Gartlgruber, A. Greco, E. Chomsky, T. G. P. Grünewald, K.-O. Henrich, A. Tanay, C. Herrmann, T. Höfer, F. Westermann
Hartlieb SA, Sieverling L, Nadler-Holly M, Ziehm M, Toprak UH, Herrmann C, Ishaque N, Okonechnikov K, Gartlgruber M, Park YG, Wecht EM, Savelyeva L, Henrich KO, Rosswog C, Fischer M, Hero B, Jones DTW, Pfaff E, Witt O, Pfister SM, Volckmann R, Koster J, Kiesel K, Rippe K, Taschner-Mandl S, Ambros P, Brors B, Selbach M, Feuerbach L, Westermann F
Gartlgruber M, Sharma AK, Quintero A, Dreidax D, Jansky S, Park Y-G, Kreth S, Meder J, Doncevic D, Saary P, Toprak UH, Ishaque N, Afanasyeva E, Wecht E, Koster J, Versteeg R, Grünewald TGP, Jones DTW, Pfister SM, Henrich K-O, van Nes J, Herrmann C*, Westermann F*
Get in touch with us


