Whole Genome Genotyping
Whole Genome Genotyping
We perform whole genome genotyping assays, using Illumina’s Infinium assay technology. This allows large-scale interrogation of variations in the human genome, at very high call rates (routinely >99%).
The Infinium Assay protocol features single-tube sample preparation without PCR or ligation steps, significantly reducing sample handling errors. An enzymatic discrimination step provides high call rates and accuracy with unambigous locus selection. We can scale the Infinium assays by parallel handling of multiple samples at a time without compromising data quality. Infinium makes significant use of tag SNPs.
Selection of supported Infinium genotyping assays:
- Infinium Core-24
- Infinium OncoArray-500K
- Infinium Global Diversity Array with Cytogenetics-8 (GDAcyto)
- Infinium OmniExpress-24
- Infinium Omni5-Quad
…and many more. Please enquire for other BeadChips.
The individual BeadChips have different minimum sample numbers. Please enquire while planning your project also to define the type of array required for your research by contacting us directly.
YOU supply
- Completion of sample sheet
- DNA from fresh frozen material: >0,5 µg DNA with av. fragment size >3kb (free of organic solvents) at a minimum concentration of 50ng/µl
- DNA from FFPE material: > 500ng at a minimum concentration of 12,5ng/µl
- Qubit or Picogreen measured DNA concentration is preferred for both sample types
- to minimize the lost of sample material during QC adjust the sample volume to 12µl for both sample types
- All samples must be submitted in a 96-well plate (e.g. 96-well FrameStar (q-)PCR Plate, skirted, black grid, SL-AM0960, Steinbrenner)
WE perform
- QC of DNA sample (applied methods depend on the DNA's origin)
- Restore procedure (only for FFPE samples)
- Infinium assay (technical reproducibility >0,98, PCR free assay)
- Basic data analysis
WE provide
- Raw data
- Genotyping information on all SNPs tested (in MS Excel 2007 compatible format)
- Medium term storage of all data for at least 3 months
- Discussion on the results
Pricing per DNA sample is provided on our pricelist. For further details concerning genotyping analysis, please contact Dr. Melanie Bewerunge-Hudler.
Infinium Technology
The Infinium Beadchips have several advantages:
- Average 15-30-fold redundancy for each SNP
- Call rates >99%
- Reproducibility >99.9%
- Most BeadArrays carry a "gene-centered" SNP selection (tag SNPs)
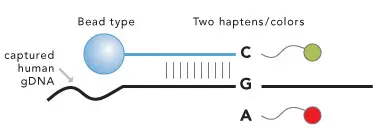
The Infinium technology is a multi step method to identify allele variations at many loci within the genome in parallel. It combines single tube sample preparation with virtualy unlimited assay multiplex scalability, dependent only on the number of features on the array.
- Whole Genome Amplification (WGA) of genomic DNA
- Fragmentation of DNA
- Precipitation and Resuspension
- Hybridization of Samples onto Beadchip
- Primer Extention of allele-specific Oligonucleotides (s. fig. 1)
- Scanning of BeadChip
- Data Analysis