FFPE Samples
Expression Profiling using FFPE Samples
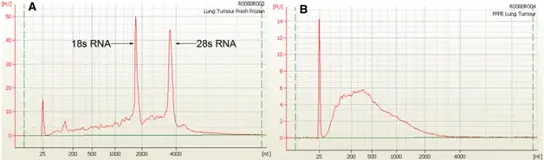
The majority of expression profiling publications use high quality RNA from frozen samples. However these studies have been restricted due to the small number of samples in these collections. On the other hand, there is a huge resource of FFPE (formalin-fixed and paraffin embedded) tissues specimens held in histopathology departments. These samples provide an invaluable resource for retrospective studies correlating molecular features with therapeutic response and clinical outcome. However extraction of RNA from these tissues has proved to be problematic due to the detrimental effects of formalin-fixation (s. fig. Comparison of RNA Quality).
We have implemented Affymetrix’s GeneChip WT Pico Kit (for GeneChip® Whole-Transcript WT) Expression Arrays) and the GeneChip 3´IVT Pico Kit (for GeneChip® 3' Expression Arrays) using RNA isolated from FFPE samples.
YOU supply
- Completion of sample sheet
- Amount of RNA >100-500 ng, to allow for input QC
- Concentration of >10 ng/µl
- Average RNA fragment size needs to be similar for all samples to be compared
- Indicate "FFPE samples" in the Remarks field for sample information in the submission form
- We recommend to run at least twice as many biological replicates as compared to classical expression profiling done by using intact RNA samples
All samples should be treated with DNase.
Samples must be non-pathogenic and non-infectious (S1-Condition).
RNA samples must be provided in a 1.5ml-tube and on dry-ice
Please consult the pricelist for detailed cost information.
We perform
- Incoming QC for quality and concentration of all samples (Nanodrop ND-1000, Agilent 2100 Bioanalyzer)
- Labeling and hybridization to the Affymetrix microarrays you request
- Monitoring the quality at all steps
- Basic Data Analysis
Output to YOU
- All RNA and labeling QC data (e.g. conc. and quality of input RNA) on request
- Raw data of microarrays
- Individual chip QC (for chip integrity and hybridization success)
- Raw data evaluation (box plots)
- Interpreted data (numerical readout and gene annotation for microarray elements (probe sets)
- Normalization across each chip
- Normalization (if wanted) for complete sets of experiments
- Group Comparisons
What's next?
You may unlock the insights buried in your experimental data, applying statistical analysis using DKFZ' in-house Chipster installation that makes use of R and Bioconductor packages and a Java GUI. Pathway analysis tools like Ingenuity Pathway Analysis or DAVID are also available via direct links, or provision of a computer workstation at GPCF (booking system for Ingenuity). Check out our webpage for more details and to get access.
For more Information, please review the FAQ site on expression profiling and the respective glossary.
Citing our service
Please cite our service in the acknowledgements part of your paper in case we've just performed the standard service. The following statement may be used.
"We thank the microarray unit of the DKFZ Genomics and Proteomics Core Facility for providing the Affymetrix Expression Profiling service."