Research
← Back to Immunotherapy and Immunoprevention
Cancer immunotherapies aim to make use of the two hallmarks of the immune system, specificity of antigen recognition and development of immunological memory. Thus, immunotherapies could be designed to only affect tumor cells and no healthy tissue, and they could also be effective against future metastases. Among different immunotherapy approaches, therapeutic cancer vaccinations aim at activating a patient’s immune system so that it eradicates an existing tumor or cancer precursor lesions. To this end, the tumor cells must have characteristics that allow the immune system to differentiate them from healthy cells. These so-called tumor-specific antigens can either be of viral origin or derive from tumor-specific mutations. We investigate both classes of tumor-specific antigens within the group.
Research Focus 1: Development of a therapeutic human papillomavirus (HPV) vaccine

Cervical carcinoma and several other malignancies arise as a result of persistent infection with high-risk types of human papillomavirus (HPV). Natural history studies indicate that nearly every sexually active individual will acquire at least one high-risk HPV infection during their lifetime. Fortunately, most infections are cleared by the immune system within 1-2 years of acquisition. Persistent infection only develops in about 2% of high-risk HPV infected people. The overall aim of this research focus is the development of a therapeutic HPV vaccine, to induce immune-mediated HPV clearance also in these patients. This is in line with current WHO efforts, which defined preferred product characteristics (PPCs) for therapeutic HPV vaccines in 2024.
Target epitope identification and validation
Cytotoxic T cells kill infected or virally transformed cells after recognizing bits of viral proteins, so-called epitopes, which are presented on human leukocyte antigen (HLA) molecules on the cell surface. There are thousands of different HLA types, all presenting different epitopes. As every human being has a different set of HLA molecules, epitopes for all major HLA groups need to be defined to generate a therapeutic cancer vaccine that is applicable to everyone regardless of the person’s HLA type.
We are currently working on the precise identification which HPV epitopes are presented on HPV-transformed tumor cells. This is important, as not every possible HPV epitope is visible to the immune system due to viral immune evasion mechanisms. For epitope identification, we predict possible epitopes in silico, confirm binding experimentally in competition-based cellular binding assays, and assess epitope surface presentation by highly sensitive mass spectrometry-based immunopeptidomics approaches. Detected epitopes are further tested for immunogenicity and for their potential to mediate T cell-cytotoxicity against tumor cells. Only epitopes that are presented to the immune system on target cells, are found reproducibly on several tumor samples, and have been proven to elicit cytotoxic immune responses are considered valid candidates for inclusion into a vaccine.
Vaccine formulation
To elicit immune responses active against tumors, it is important to induce high numbers of tumor-specific cytotoxic T cells (CTLs). Strong CTL responses and formation of immunological memory need the contribution of T helper cells, therefore we always include T helper epitopes in our vaccine formulations. Adjuvants and mode of delivery are also important aspects of vaccine design. Epitopes can be administered as peptides, but also e.g. encoded in RNA constructs, or included into nanoparticle formulations. Various strategies are explored to determine the best way of delivery for inducing strong anti-HPV immune responses. Moreover, we have developed tumor models that allow us to assess the anti-tumor efficacy of our vaccines.
Directing tumor cells to the tumor site
Many tumors have evolved ways of preventing T cell infiltration, or of making intratumoral T cells functionally inactive. We therefore explore strategies of improving the trafficking of vaccination-induced T cells to the tumor site. To counteract immunosuppressive mechanisms in the tumor microenvironment, we investigate local and systemic immunotherapy combination approaches. In order to enhance HPV-derived epitope presentation and reshape the tumor immune microenvironment to a more immunoresponsive phenotype, current combination strategies investigated in the lab include low dose irradiation as well as Proteolysis Targeting Chimeras (PROTACs).
Research Focus 2: Identification & validation of mutation-derived tumor neoepitopes as immunotherapy targets
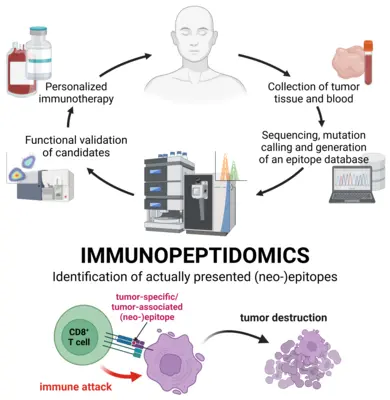
The interest in mutation-derived tumor neoepitopes as immunotherapy targets has been prompted by the clinical success seen with checkpoint blockade therapies. It was found that effective immune responses triggered by these therapies were directed against neoepitopes. Thus, therapies specifically targeting these epitopes, such as neoepitope-specific vaccinations or adoptive transfer of neoepitope-specific T cells, could combine the efficacy of existing immunotherapeutic approaches with absolute tumor-specificity.
The current workflows for neoepitope detection combine tumor genome sequencing, identification of mutations, assessing if these mutations are translated into proteins, and epitope prediction for the patient’s HLA type. The next step commonly is testing for T cell reactivity against predicted neoepitopes. However, this does not provide an answer to the question if a given neoepitope is truly present on the surface of the tumor cells, and thus represents a valid immunotherapy target.
The high-sensitivity targeted immunopeptidomics methodology that we have developed for identification of HPV epitopes can also be used for detection of other hard-to-detect epitopes. However this workflow is limited to an a priori defined set of analytes and typically not capable to assess all potential candidates of interest generated by a patient’s mutational spectrum. Thus, more recently we have established an untargeted immunopeptidomics workflow allowing us to generate a comprehensive overview of a tumor’s immunopeptidome at reduced depth. The two workflows are highly complementary and we have been able to show that both of them are able to detect distinct sets of mutation-derived, tumor-specific neoepitopes.
Therefore, we have set up a consortium of groups interested in neoepitope detection in the scope of the National Center for Tumor Diseases (NCT) Heidelberg, providing expertise in tumor sequencing, mutation calling, neoepitope prediction and immunological assessment facilities. Within this consortium our group provides the immunopeptidomics expertise in order to proof actual HLA-mediated surface presentation of neoepitopes to complete the neoepitope detection pipeline. We act has the Immunopeptidomics Unit of the NCT. In the future we aim to fully integrate our workflows into the clinical routine to provide validated targets for every patient and contribute to the development of highly specific future immunotherapies.
Besides our interest in clinical application of immunopeptidomics, we continue to develop new and improved methodologies. We apply immunopeptidomics in the pre-clinical setting to e.g. investigate the effects of different drugs on the immunopeptidome, characterize prominent antigens for their presented epitopes to facilitate off-the-shelf immunotherapies and to assess the antigen presentation potential of vaccines candidates in vitro.
← Back to Immunotherapy and Immunoprevention