Immune Diversity
- Immunology, Infection and Cancer

Prof. Dr. Nina Papavasiliou
Abteilungsleitung
Unlike most other cell types in the body, which are static, immune cells must travel through (and adapt to) multiple different environments on a constant basis. This ability to adapt is predicated on molecular mechanisms that generate informational diversity. Overall, the goal of our research is to understand the molecular processes that generate phenotypic diversity, which in turn is required for adaptation.
Our Research
Unlike most other cell types in the body, which are static, immune cells must travel through (and adapt to) multiple different environments on a constant basis. This ability to adapt is predicated on molecular mechanisms that generate informational diversity, such that every immune cell is genetically different than every other (which is the case with cells of the adaptive immune system, such as B and T lymphocytes) or transcriptomically different than their siblings (which appears to be the case with innate immune cells, like macrophages). Overall, the goal of our research is to understand the molecular processes that generate phenotypic diversity, which in turn is required for adaptation.
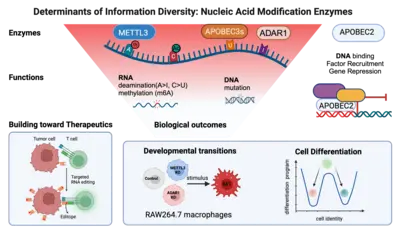
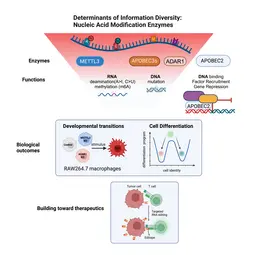
Of the large set of enzymes known to modify RNA or DNA bases through addition (or removal) of specific chemical groups, we have chosen three classes.
(1) We are interested in AID/APOBEC proteins that deaminate cytosine (C) to uridine (U) in RNA (or DNA) to yield uracil (U). Members of this family of proteins can be highly substrate specific (e.g. AID mutates only DNA); others can be rather promiscuous (e.g. APOBEC1 deaminates both RNA and DNA) and yet others (like APOBEC2) appear to only be able to bind DNA - but not catalyze deamination despite the presence of a perfectly functional catalytic site.
(2) We are curious about ADAR proteins, that deaminate adenosine (A) to inosine (I) in RNA. We would like to understand their contribution to information diversity in cells, and to learn why deregulation is linked with oncogenesis in almost all tissues tested.
(3) We are beginning to also study enzymes that methylate RNA e.g. on adenosine. We would like to understand the interplay between methylation and deamination on the same base (e.g. adenosine) and the role this plays in immune cell diversity.
In a bottom-up approach we study these enzymes from their molecular mechanism to their biological outcomes at the cellular level. For example, we study how loss of individual RNA modification enzymes affects cellular differentiation and how the overexpression of others can be causal to oncogenesis. Eventually we would like to understand how an ensemble of modifications on a single transcript defines its fate (its location, half life and translation efficiency) and we are collaborating with mathematicians to develop novel algorithms toward that. Finally, we keep a keen eye on translation: for example (a) we are using guided RNA editing (through endogenous ADAR engagement) to convert HLA-presented epitopes to neoepitopes (or "editopes") to potentiate immune recognition e.g. of tumours by T cells and (b) co-developing novel fusogenic delivery particles to deliver such cargo to cells of interest.
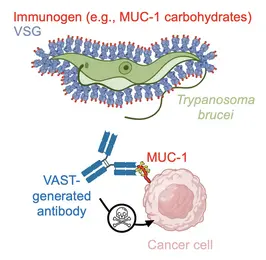
We also study diversity at the protein level. For that we use a model organism (the African trypanosome) that continually "resurfaces" its dense protein coat to evade the antibody response. We would like to understand the biophysical features of the coat (which is composed by 11 million repeats of a single "Variant Surface Glycoprotein" or VSG) that allow the elicitation of an acutely focused immune response, which then the organism evades by effectively "resurfacing" the coat (through replacement of entire VSGs or with parts thereof). Toward this we use antibody repertoire analyses as well as structural means. We aim toward a mechanistic understanding of a changing antibody response toward an antigenic coat that changes just as rapidly, and their co-evolution through time. At the same time, we are utilizing early knowledge from this system to generate better immunogens (VAST) with strong translational potential.
Teams
Team “RNA Editing and Modifications”
Team leader: Dr. Riccardo Pecori
Team members: Annette Arnold, Dr. Beatrice Casati, Dr. Salvatore Di Giorgio, Valerie Griesche, Eliana Patricelli, Laura Pezzella
Research Summary
RNA molecules undergo various processing events that diversify genomic information, including single-base modifications. Dysregulation of these processes has been linked to multiple diseases.
Our team “RNA Editing and Modifications” studies these events. Our current focus is on RNA deamination by ADAR1 and APOBEC1 enzymes, as well as RNA methylation by METTL3. In our work we explore the role of these enzymes (and the modifications they catalyze) in health and disease, with particular focus on the immune system.
For example, we study how adenosine deamination affects developmental transitions of macrophages, and how loss of ADAR1 might impact adenosine methylation (m6A levels). We study this at the sequence level (using novel nanopore sequencing pipelines in collaboration with Terry Lyons at Oxford and Tessy Papavasiliou at Warwick), at the transcript processing level (in collaboration with Georg Stoecklin at Heidelberg University) and at the level of the cell and the decisions it can make that rely on proper "reading" of these modifications.
We also study ADAR1-mediated deamination and specifically, how ADAR1 can be recruited by specialized guide RNAs (ADAR1 engagers) to generate changes in specific transcripts of interest to us. A current example is the implementation of novel precision tools to recruit endogenous ADAR1 with high efficiency to regions of transcripts that encode HLA presented peptides, and to alter those peptides in a way that makes them immunogenic (i.e. "discoverable" by T cells). There is substantial therapeutic potential in this approach which we are taking from the mechanistic level (generation of novel guide RNAs) to the organismal level (preclinical work on tumour rejection after procurement of ADAR1 engagers).
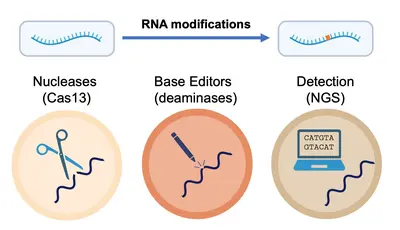
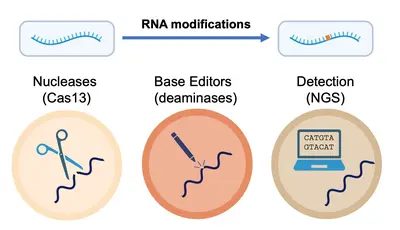
More broadly, we take knowledge gained from studying RNA modifications in the context of the cell and apply it to develop cutting-edge technologies for the precise detection and manipulation of RNA at the single-nucleotide level. In tandem, we develop collaboratively and implement advanced strategies for the rapid and accurate detection and quantification of RNA modifications across diverse data types, thus driving a deeper understanding of oncogenesis and immune responses, but also paving the way for novel therapeutic strategies.
Grants
- 2022, DFG GRK2727 "ImCheck" (project A1.2)
- 2022, Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) project number 439669440 TRR319 RMaP TP 04 awarded to F.N.P.
- 2021, HI-TRON Kick-Start Seed Funding Program 2021 awarded to Dr. Pecori and Prof. Dr. Tenzer (University Medical Center Mainz).
- 2020, Ministry of Science, Research and the Arts of Baden-Württemberg for COVID-19 research (grant agreement no. Kap. 1499 TG 93 to Dr. Pecori, Prof. Dr. Papavasiliou and Prof. Dr. Miethke (Medical Faculty of Mannheim)).
- 2020, European Research Council award (ERC PoC "EpiMODkit") to Prof. Dr. Papavasiliou.
Collaborations
- Prof. Dr. med. Michael Platten (DKFZ)
- Prof. Dr. Rienk Offringa (DKFZ)
- Dr. Rafael Carretero (DKFZ)
- Prof. Dr. Mark Helm (Uni Mainz)
- Prof. Dr. Rocio Sotillo (DKFZ)
- Prof. Dr. Georg Stoecklin (Uni Heidelberg)
- Prof. Dr. Stefan Tenzer (University Medical Center Mainz)
- Prof. Dr. Terry Lyons (University of Oxford)
- Prof. Dr. Tessy Papavasiliou (University of Warwick)
- Prof. Dr. Thomas Miethke (Medical Faculty of Mannheim)
- Prof. Dr. Thorsten Stafforst (University Hospital Tübingen)
- Prof. Dr. Qiang Pan-Hammarström (Karolinska Institutet)
Publications
- Latifi, et al. Precise and efficient C-to-U RNA base editing with SNAP-CDAR-S. Nucleic Acids Research, 2023.
- Casati, et al. Tumor warm-up: RNA editing boosts tumor immunogenicity. BioRxiv, 2023.
- Pecori, et al. ADAR1-mediated RNA editing promotes B cell lymphomagenesis. Science, 2023.
- Pecori, et al. ADAR RNA editing on antisense RNAs results in apparent U-to-C base changes on overlapping sense transcripts. Front. Cell Dev. Biol., 2023.
- Casati, et al. Rapid, adaptable and sensitive Cas13-based COVID-19 diagnostics using ADESSO. Nature Communications, 2022.
- Pecori, et al. Functions and consequences of AID/APOBEC-mediated DNA and RNA deamination. Review, Nature Reviews Genetics, 2022.
- Stroppel, et al. Harnessing self-labeling enzymes for selective and concurrent A-to-I and C-to-U RNA base editing. Nucleic Acids Research, 2021.
- Kluesner, et al. MultiEditR: The first tool for the detection and quantification of RNA editing from Sanger sequencing demonstrates comparable fidelity to RNA-seq. Molecular Therapy Nucleic Acids, 2021.
- Casati, et al. ADAR-mediated RNA editing and its therapeutic potentials. RNA Technologies, Book Chapter, Springer Series, Vol. 11, 2021.
- Lerner, et al. C-to- U RNA editing: from computational detection to experimental validation. Book Chapter, Methods in Molecular Biology, 2021.
- Pecori, Papavasiliou. It takes two (and some distance) to tango: how ADARs join to edit RNA. News & Views, Nature Structural Molecular Biology, 2020.
- Tasakis, et al. ADAR1 can drive Multiple Myeloma progression by acting both as an RNA editor of specific transcripts and as a DNA mutator of their cognate genes. BioRxiv, 2020.
- Lerner, et al. RNA Editors, Cofactors, and mRNA Targets: An Overview of the C-to-U RNA Editing Machinery and Its Implication in Human Disease. Review, Genes, 2019.
Team “Deaminases and lymphocyte fate determination”
Team leader: Dr. Elias Amro
Team members: Christos Gkougkousis, Nina Lemmen
Research Summary
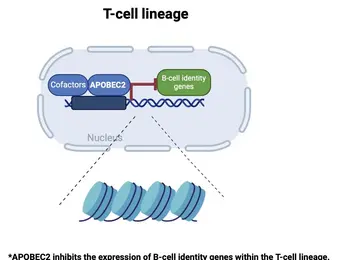
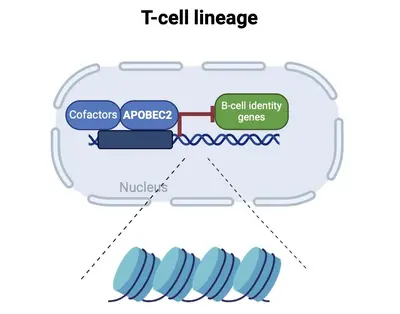
Differentiation into specific cell fates and maintenance of cell identity require molecular programs driven by transcription factors that not only sustain cell-specific identities but also suppress alternative cell fates. Our recent research has focused on exploring the role of APOBEC2 in lymphoid lineage specification. APOBEC2 is part of the highly conserved apolipoprotein B mRNA-editing enzyme catalytic polypeptide-like (APOBEC) family, known for its zinc-dependent deaminase activity (Pecori et al., 2022; Wedekind et al., 2003; Harris et al., 2002; Powell et al., 2014). While most members of this family catalyze the conversion of cytidine to uridine within DNA or RNA, resulting in C-to-U transitions, APOBEC2 lacks this enzymatic function. Despite this, APOBEC2 plays an unconventional role in regulating biological functions both in vitro and in vivo. In mice, depletion of Apobec2 leads to muscle weakness, reduced muscle mass, and a shift from fast to slow twitching muscle fiber formation (Sato et al., 2010; Mikl et al., 2005; Sato et al., 2018). Apobec2 has also been implicated in cancer development through mutations and altered gene expression (Okuyama et al., 2012; Wei et al., 2024; Li et al., 2019). Recent research from our lab (Lorenzo, Molla, Amro et al., 2024) has uncovered a novel role for APOBEC2 as a transcription factor that maintains muscle identity by suppressing non-muscle genes. Beyond muscle biology, our current work reveals that APOBEC2 deficiency in mice and humans causes defects in lymphoid lineage specification. Human individuals with homozygous APOBEC2 mutations show weakened immune responses to vaccination and have increased susceptibility to immune dysfunction (Amro, Gkougkousis et al., unpublished). To further unravel the role and function of APOBEC2 in health and disease, our team's research focuses on two main objectives:
understanding the molecular mechanisms by which Apobec2 regulates lymphoid lineage specification;
elucidating the impact of Apobec2 mutations on the human immune system functions.
Collaborations
- Dr. Charles Imbusch (Uni Mainz)
- Prof. Dr. Christoph Dieterich (Uni Heidelberg)
- Prof. Dr. Lennart Hammarström (Karolinska Institutet)
- Prof. Dr. Qian Pan Hammarström (Karolinska Institutet)
- Prof. Dr. Fowzan AlKuraya (King Faisal Hospital, Riyadh)
Publications
- Lorenzo, Molla, Amro, et al. APOBEC2 safeguards skeletal muscle cell fate through binding chromatin and regulating transcription of non-muscle genes during myoblast differentiation. Proceedings of the National Academy of Science, 2024.
- Pecori, et al. Functions and consequences of AID/APOBEC-mediated DNA and RNA deamination. Nature Reviews Genetics, 2022.
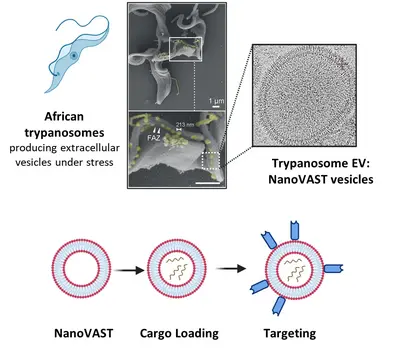
Team “T. brucei derived tools: NanoVAST”
Team leader: Dr. Katerina Spyridopoulou
Team members: Laura Pezzella, Niklas Veocic
Research Summary
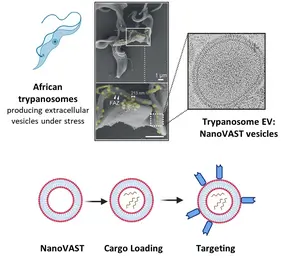
Targeted delivery of therapeutic agents to specific cell types remains a significant challenge in advancing fields such as gene therapy, RNA modification, and drug delivery. While technologies like CRISPR/Cas hold immense therapeutic potential, their clinical translation is hampered primarily by the lack of safe, efficient, and targeted delivery systems. Current delivery approaches, including electroporation and viral vectors, suffer from limitations like toxicity and immunogenicity.
To address this, our team utilizes nanoVAST, a novel platform developed by our colleagues at Panosome GmbH. NanoVAST comprises a patented vesicular phospholipid bilayer based on scalable production of African trypanosome-derived extracellular nanovesicles. Being biogenic, nanoVAST does not exhibit toxicity, while its fusogenic properties enable cytoplasmic delivery of its cargo. Critically, it is highly targetable, as its surface can be modified using a specific sortase-based chemistry – termed VAST – to attach targeting moieties.
We are optimizing nanoVAST for in vitro, ex vivo, and in vivo therapeutic cargo delivery, exploring its biochemical properties to develop highly specific cell-targeting strategies. Our work focuses on the development of cargo-specific loading methodologies and cell-specific targeting approaches for cancer and immune cells (such as T cells and NK cells). Cargo types that are currently being investigated include TCR-mRNA, CAR mRNA, siRNA, guide RNAs for RNA editing, and DNA vectors. We also develop and employ state-of-the-art technologies to assess optimized nanoVAST production, cargo loading, and effective and functional cellular delivery.
By improving the targeted delivery of drugs and gene therapy systems, this technology has the potential to unlock new therapeutic avenues for numerous diseases currently lacking effective treatments.
Grants
- European Research Council 2022 Proof of Concept Grant (nanoVAST)
- DKFZ seed funding program 2024: Kick-start DreamTeam Grant
Collaborations
- Prof. Dr. med. Michael Platten (DKFZ)
- Prof. Dr. Joachim Spatz (MPI for Medical Research)
- Synthetic Immunology Research Network
Publications
- Hempelmann, et al. Nanobody-mediated macromolecular crowding induces membrane fission and remodeling in the African trypanosome. Cell Reports, 2021.
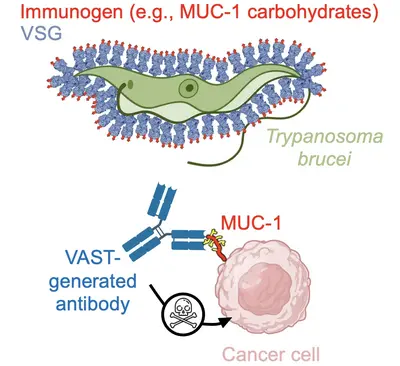
Team “T. brucei derived tools: VAST and the development of antibodies as therapeutics”
Team leader: Dr. Joseph (Joey) Peter Verdi
Team members: Dr. Monica Chandra, Dr. Anastasia Gkeka, Dr. Nataliia Kashpur, Eblina Kelmendi, Seneca Kinn-Gurzo, Dr. Jose Paulo Lorenzo, Dr. Colin Miller, Eleftheria Papamanoglu, Sandra Ruf, Dr. Eirini Sidiropoulou
Research Summary
Keytruda, Humira, Herceptin… Many human diseases can now be treated successfully with antibody-based therapies. Our team aims to expand the repertoire of antibody-treatable diseases using our internally developed technology platform for antibody generation. While it has now become relatively simplistic to generate high-quality antibodies against protein antigens, there remains a general inability to reproducibly generate the same quality of antibodies against small molecules and carbohydrates. These notably small-sized molecules are associated with or the cause of just as many diseases as proteins are, yet this space has yet to be convincingly explored. Here, we do exactly that - we specifically chase after these “difficult targets” with the intention of developing novel therapies.
To attack these difficult targets, we developed a platform technology based on the unique surface architecture of the African trypanosome. The surface of Trypanosoma brucei cells is extremely densely packed with 10 million copies of one protein - the variant surface glycoprotein (VSG). This uniquely dense and uniformly oriented array of antigens stimulates the immune system in a remarkable way, directing virtually the entire disease-relevant antibody response towards epitopes localised on just the very top of the VSG protein. This so-called “epitope focusing” effect is perhaps the strongest of its kind in nature, which our team has harnessed by “sortagging” (fusing) these difficult targets to the surface of VSG and creating the VSG Immunogen Array by Sortase Tagging - the VAST. As examples of the VAST’s applicability, the technology has so far been used to make a series of pico- and femtomolar affinity antibodies to the opioids fentanyl and carfentanil that can completely prevent opioid toxicity in animal model systems, as well as a series of high-quality antibodies to carbohydrates present on mucin 1 (MUC1), a high-priority target in solid tumor cancers. This exciting technology, its antibody products, and every exceptionally talented member of this team form the cornerstone of a biotechnology start-up company, Panosome GmbH, located in the local Heidelberg area.
Grants
- 2024, KMU-innovativ (BMBF) project “CarboVAST” awarded to Panosome GmbH with DKFZ as a collaborating institution (Dr. Verdi and Prof. Papavasiliou as project leaders)
- 2024, KMU-innovativ (BMBF) project “GliomAb” awarded to Panosome GmbH with DKFZ as a collaborating institution (Dr. Verdi and Prof. Papavasiliou as project leaders).
- European Research Council 2020 Proof of Concept Grant (EpiMODkit)
Collaborations
- Prof. Dr. Rienk Offringa (DKFZ)
- Prof. Dr. med. Michael Platten (DKFZ)
- Dr. Aubry Miller (DKFZ)
- Dr. Erec Stebbins (DKFZ)
Selected Publications
- Urban and Gkeka et al., The fentanyl-specific antibody FenAb024 can shield against carfentanil effects. Toxicology Letters, 2024.
- Triller et al., A trypanosome-derived immunotherapeutics platform elicits potent high-affinity antibodies, negating the effects of the synthetic opioid fentanyl. Cell Reports, 2023.
- Gkeka and Aresta-Branco et al., Immunodominant surface epitopes power immune evasion in the African trypanosome. Cell Reports, 2023.
- Triller et al., Immunization with Genetically Modified Trypanosomes Provides Protection against Transmissible Spongiform Encephalopathies. International Journal of Molecular Sciences, 2022.
Lab members
-

Prof. Dr. Nina Papavasiliou
Head of Division
-

Dr. Elias Amro
Team leader "Deaminases and lymphocyte fate determination"
-
Annette Arnold
Technische Assistent
-
Dr. Monica Chandra
Postdoctoral researcher
-
Dr. Salvatore Di Giorgio
Postdoctoral researcher
-
Anastasia Gkeka
Postdoctoral researcher
-
Christos Gkougkousis
Doctoral researcher
-

Valerie Griesche
Doctoral researcher
-
Nataliia Kashpur
-
Seneca Kinn-Gurzo
Doctoral researcher
-
Dr. Jose Paulo Lorenzo
Postdoctoral researcher
-
Eleftheria Papamanoglou
-

Dr. Riccardo Pecori
Deputy head of division and team leader "RNA editing and modifications"
-
Laura Pezzella
Doctoral researcher
-
Sandra Ruf
-
Dr. Eirini Sidiropoulou
Postdoctoral researcher
-

Dr. Aikaterini Spyridopoulou
Team leader "T. brucei derived tools: NanoVAST"
-
Dr. Joseph Peter Verdi
Team leader "T. brucei derived tools: VAST and the development of antibodies as therapeutics"
- Show profile

Christoph Tim Pufall
Administrative Assistant
-

Colin Gregory Miller
Postdoctoral researcher
-
Dr. Katharina Urban
-

Eblina Boddin
Alumni
In Science, people come and go. This is also part of research. Several people have also worked together in our team since the foundation of our research group in 2016. They all contributed to making important progress in our research projects. Here is a list of all the people that worked with us (in brackets their previous role within our group).
Selected Publications
Lorenzo JP, Molla L, Amro EM, Ibarra IL, Ruf S, Neber C, Gkougkousis C, Ridani J, Subramani PG, Boulais J, Harjanto D, Vonica A, Di Noia JM, Dieterich C, Zaugg JB, Papavasiliou FN.
Pecori R, Rent W, Pirmoradian M, Wang X, Liu D, Berglund M, Li W, Tasakis RN, Di Giorgio S, Ye X, Li X, Arnold A, Wüst S, Schneider M, Selvasaravanan K, Fuell Y, Stafforst T, Amini RM, Sonnevi K, Enblad G, Sander B, Wahlin BE, Wu K, Zhang H, Helm D, Binder M, Papavasiliou FN.
Gkeka A, Aresta-Branco F, Triller G, Vlachou EP, van Straaten M, Lilic M, Olinares PDB, Perez K, Chait BT, Blatnik R, Ruppert T, Verdi JP, Stebbins CE, Papavasiliou FN.
Triller G, Vlachou EP, Hashemi H, van Straaten M, Zeelen JP, Kelemen Y, Baehr C, Marker CL, Ruf S, Svirina A, Chandra M, Urban K, Gkeka A, Kruse S, Baumann A, Miller AK, Bartel M, Pravetoni M, Stebbins CE, Papavasiliou FN, Verdi JP.
Casati B, Verdi JP, Hempelmann A, Kittel M, Gutierrez Klaebisch A, Meister B, Welker S, Asthana S, Di Giorgio S, Boskovic P, Man KH, Schopp M, Ginno PA, Radlwimmer B, Stebbins CE, Miethke T, Papavasiliou FN, Pecori R.
Pecori R, Di Giorgio S, Lorenzo JP, Papavasiliou FN.
Hempelmann A, Hartleb L, van Straaten M, Hashemi H, Zeelen JP, Bongers K, Papavasiliou FN, Engstler M, Stebbins CE, Jones NG.
Casati B, Stamkopoulou D, Tasakis RN, Pecori R.
Kluesner M, Tasakis RN, Lerner T, Arnold A, Wüst S, Binder M, Webber BR, Moriarity BS, Pecori R.
Get in touch with us