Epithelium Microbiome lnteractions
- Immunology, Infection and Cancer
- Junior Research Group

Dr. Jens Puschhof
Head of Division
In the EMIL group, we study how bacteria contribute to cancer development, metastasis and treatment response. Our team includes cancer researchers, tissue engineers, microbiologists and bioinformaticians working together to help patients by deciphering the functional impacts of bacteria in human tumors.

Our Research
In the EMIL group, we study how bacteria contribute to cancer development, metastasis and treatment response. Our team includes cancer researchers, tissue engineers, microbiologists and bioinformaticians working together to help patients by deciphering the functional impacts of bacteria in human tumors.
Using tissue samples from patients, we isolate bacterial strains and create organoids. These models can be combined with additional features of the human gut on USB stick-sized devices, “organ chips”, for detailed studies. We further transplant organoid-bacteria mixtures into mouse models to study their migration and interaction in a whole body context.
In our unique biobank at DKFZ, we characterize both tumor organoids and tumor-resident bacterial strains through whole genome sequencing, fluorescence microscopy and other cutting-edge methods. Based on the diverse data obtained, we develop hypotheses about how specific bacterial species might influence tumors. With our broad portfolio of patient-derived models and assays, our main focus lies on building a deep understanding of these relationships. Through close collaborations with clinical partners, we work towards the rapid translation of our findings towards new microbiome-based detection, prevention and treatment strategies of cancer.
If you like working in or collaborating with a young team with flat hierarchies, a collaborative spirit and a keen interest in functional cancer microenvironment work, please get in touch!
Projects

In the METABAC project, we investigate how bacteria contribute to the process of metastasis. Using patient-derived organoids, organ chips and organoid xenotransplantations, we study how specific bacterial strains shape the multiple steps of the metastatic cascade.

We are coordinating the BACTORG consortium (with Melanie Börries from Freiburg) to yield translational breakthroughs in cancer microbiome science in cooperation with 7 leading medical centers in Germany and the Netherlands. Using patient-derived bacterial strains and organoids, we validate the functional relevance of clinical multi-omics datasets. In doing so, we envision to design personalized prevention and treatment strategies for colorectal cancer that take the gut and tumor microbiome into account.

In collaboration with several international partners, we are exploring the role of the microbiome in liver cancer development, progression and treatment response. Using patient-derived organoids of different types of liver cancer and tumor-relevant bacterial strains, we perform diverse phenotypic assays to elucidate functional roles of these bacteria for subsets of liver cancer patients.
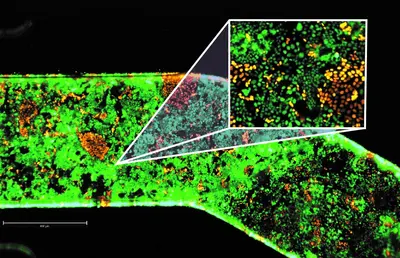
In collaboration with Emulate Inc, we develop next-generation organ chip platforms that allow us to study the impacts of bacteria on colorectal cancer migration and metastasis formation. Using diverse patient-derived organoids and further microenvironmental components, we re-build the complexity of the tumor microenvironment one step at a time.

Projects in our group are meant to generate groundbreaking basic scientific insights and translate those to patient benefit in cancer prevention and treatment. Your project with us should allow you to follow your own scientific interests in collaboration with the team and boost your scientific profile. Therefore, your project will emerge from the intersection of your experiences, future plans, and what our lab environment can enable you to do. Most PhD and postdoc projects in our group can be summarized in 2 categories:
1. Functional studies on bacterial strains in cancer development and progression.
These include organoid and organ chip culturing, bacterial culturing, genetic engineering with CRISPR Cas9, co-culture development, immune cell co-cultures and mouse model transplantations.
2. Integrative bioinformatics of host and microbiome in patients.
In this type of project, you can do up to 80% dry lab work, coupled to generating your own datasets and validating the patterns you discover together with group members and external collaborators. Our focus is on host transcriptomics (single cell and bulk), bacterial strain whole genome sequencing and microbiome-targeted sequencing of patient tissues.
If you like working in a young team with flat hierarchies, a collaborative spirit and a keen interest in functional cancer microenvironment work, get in touch!
Team
-

Dr. Jens Puschhof
Head of Division
-

Jarryd Michael Boath
Postdoc
-

Anna Boot
Master Student
-

Aysegül Durgun
Master Student
-

Paula Feldt
Master Student
-

Clara Foerster
Bachelor Student
-
Nimisha Khurana
-
Despoina Evgenia Kiousi
Visiting Postdoc
-

Kamil Moskal
PhD Student
-

Luigi Lukas Nardella
PhD Student
-

Kyanna Ouyang
PhD Student
-
Chrysanthi Ionna Psarologaki
-

Darline Rosel Dagmar Rößner
Student Assistant
-

Lena Schorr
PhD Student
-

Mahero Ayesha Shah
Technical Assistant
-

Nina Stinchcombe
PhD Student
-

Julia Tabel
Master Student
-

Dr. Smriti Verma
Staff Scientist
Selected Publications
Modeling cancer-microbiome interactions in vitro – a guide to co-culture platforms.
Moskal K, Khurana N*, Siegert L*, Lee YS*, Clevers H, Elinav E, Puschhof J.
Sticky situation as bacteria drive colon cancer.
Ouyang KS, Puschhof J.
Pleguezuelos-Manzano C*, Puschhof J*, Rosendahl Huber A*, et al.
Puschhof J*, Pleguezuelos-Manzano C*, Martinez-Silgado A, Akkerman N, Saftien A, Boot C, de Waal A, Beumer J, Dutta D, Heo I, Clevers H
Rosendahl Huber A*, Pleguezuelos-Manzano C*, Puschhof J*, Ubels J*, Boot C, Saftien A, Verheul M, Trabut LT, Groenen N, van Roosmalen M, Ouyang KS, Wood H, Quirke P, Meijer G, Cuppen E, Clevers H, van Boxtel R
Schorr L*, Mathies M*, Elinav E and Puschhof J
Get in touch with us

