Federated Information Systems
- Imaging and Radiooncology
- DKFZ Hector Cancer Institute

Prof. Dr. Martin Lablans
Head of Department
To build bridges for biomedical research, we pioneer novel methods solving both technical and medico-legal data sharing issues. We create real-world networks for joint use of data and biosamples in Europe and beyond.
Our Research
Data-driven research in the health sector depends therefore on the exploitation of distributed sources of data and tissue samples. Two significant fields of research emerge from these requirements. First, establishing the basis for carrying out research efficiently in networked federations at the technical and data content levels. Second, the implementation of regulatory requirements, which arise naturally during the processing of sensitive patient data (E.g. data protection).
In the Department of Federated Information Systems, we investigate problems that frequently arise in networked medical research:
- Semantics of data, to provide a common understanding of their meaning
- Multi-center integration of data from heterogeneous sources
- Data protection, consent management, record linkage and pseudonymization
- Distributed processes for evaluating and exchanging data
- Measurement and improvement of data quality over multiple sites
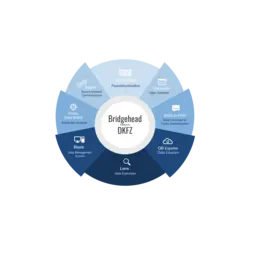
We are actively engaged in developing interoperable and reusable tools for tackling the problems in this area. In addition, we have deployed our solutions in national and international treatment federations. With our “Bridgeheads” we have created a network of exceptional partners (cf. diagram).
Projects and Networks
To build bridges for biomedical research, we pioneer novel methods solving both technical and medico-legal data sharing issues. We create real-world networks for joint use of data and biosamples in Europe and beyond.
We build bridges among institutions with GDPR-compliant federated data management solutions, including the “Mainzelliste” for pseudonymization and record linkage, several federated search solutions and the “Bridgehead” for controlled data sharing. These form the backbone of the German Cancer Consortium (DKTK), lung cancer patients data collection in the National Network Genomic Medicine (nNGM) and the “Sample Locator”, which allows federated search across 16 European biobanks (BBMRI-ERIC). As a bridge division to the University Medicine Mannheim, we foster data-driven collaborations as part of the DKFZ Hector Cancer Institute’s novel approach to data sharing with the Medical Informatics Initiative.

The German Consortium for Translational Cancer Research (DKTK) is one of six German Centres for Health Research (DZG) funded by the BMBF. The cross-site networking of the consortium is made possible by the Clinical Communication Platform (CCP-IT), which is being developed by a dedicated working group consisting of at least one member from each partner site. Thanks to the federated network architecture, data from tumour documentation and biomaterial banks can also be contributed by patients before the DKTK is founded, while maintaining data protection and data sovereignty for the consortium. Feasibility studies and the recruitment of studies are thus supported not only during, but also prior to their application to the DKTK.
Website: Clinical Communication Platform

The aim of the DKFZ-Hector Cancer Institute at the University Medical Centre Mannheim is to accelerate the transfer of results from cutting-edge oncological research into patient care (translation) and to make the use of findings from everyday clinical practice available for cancer research (reverse translation) by pooling expertise in the field of cancer research and cancer medicine. A concept for the transfer of routine data from patients at the UMM was developed in coordination with the Data Integration Center of the UMM in order to automatically transfer routine clinical data from study patients. A graphical user interface triggers the generation of patient pseudonyms at the DKFZ and enables their disclosure to a trusted third party (TTP) in the UMM hospital network (Hospital TTP). This "link" between the DKFZ and the UMM serves as the legal basis and proof for the subsequent transmission of medical patient data.

The Helmholtz Institute for Translational Oncology Mainz (HI-TRON Mainz) is to be established in the science city of Mainz as a world-leading centre for personalised cancer medicine with a focus on immunotherapy.
The HI-TRON Mainz Data Portal is being developed as a central point of the facility and to support sustainable data management. This is a data catalogue with metadata that is designed to give scientists an overview of potentially available data and biosamples.
The portal allows scientists to search for expression profiles or molecular signatures in other study and tumour entities, for example. It also enables cross-OMIC integrations to develop multidimensional classifiers using machine learning.

The HiGHmed consortium is working on novel, interoperable solutions in medical informatics with the aim of making medical patient data accessible for clinical research and teaching. The project combines and integrates the expertise of 12 leading university hospitals in Germany as well as other partners from science and industry. The oncology use case rises to the challenge of integrating enormous amounts of data from genome sequencing and radiology into clinical practice. A virtual oncology centre is to visualise the course of treatment for cancer patients and serve as an exchange platform for hospitals, research institutions, doctors and patients. This will enable similar cancer cases to be better identified and individual patient-oriented treatment to be provided.

In the future, all patients in Germany with advanced lung cancer will have access to molecular diagnostics and innovative therapies through a national network. To achieve this, 15 university cancer centres are joining forces in the National Network for Genomic Medicine (nNGM) – including all centres of the German Consortium for Translational Cancer Research (DKTK) and all oncology centres of excellence currently funded by the DKH. The basis for all distributed processes is a secure network of Bridgeheads for the Clinical Communication Platform (CCP-IT). Thanks to its expansion through the Connecting Comprehensive Cancer Centers (C4) initiative, this platform also includes all nNGM centres.

As part of the German Biobanking Alliance, a distributed team of 20 computer scientists developed an IT platform for the exchange of biosamples in order to compile large, multi-centre sample collections for research projects. Here, the biobanks are networked both within the German consortium and with international biobank infrastructures such as BBMRI-ERIC.

The German Centres for Health Research (DZG) are looking for new therapies for diabetes, infections, lung diseases, cancer, cardiovascular diseases, neurodegenerative diseases and mental illnesses. Our department represents the German Cancer Consortium (DKTK) in the DZG-wide Research IT Working Group.

The Pan-European Biobank and Biomolecular Research Infrastructure (BBMRI-ERIC) is a distributed biomedical and life science structure for the sustainable storage and distribution of biobank samples and related data in Europe. The BBMRI-Eric provides access to biobank and biomolecular resources and their expertise and services.

Cancer Core Europe is a consortium of seven leading cancer centres from Europe. It was founded in 2014 to accelerate the development of innovative cancer therapies through close collaboration in translational and clinical research. One of the four main pillars is the VDC (Virtual Data Centre), which makes this vision possible through data sharing. The bridgehead developed in our department enables data sharing between the participating sites. It started with 4 pilots (Gustave Roussy (France), Karolinska Institutet (Sweden), Vall d'Hebron Institute of Oncology (Spain) and Netherlands Cancer Institute (Netherlands). A comprehensive data protection concept, developed together with the data protection officers of the participating institutions, forms the basis for the use of patient data.

The European Cancer Imaging Initiative (EUCAIM) connects research institutions, healthcare providers and commercial innovators from currently 12 countries across Europe. EUCAIM provides an interoperable, privacy-compliant and secure infrastructure for conducting federated, distributed analysis of annotated, anonymised cancer imaging data. We are developing EUCAIM's federated exploration system based on Samply.Lens and Samply.Beam.

The aim of this project, part of the ITCC paediatric cancer data portal initiative, is to establish a sustainable solution for the systematic prioritisation of new cancer therapies for children in Europe and worldwide by making better use of existing molecular and clinical data, thus offering new hope to children with previously incurable cancers. Metadata harmonisation and data linking are essential to achieve this. The Hopp-ITCC International Data Integration Platform will help to prioritise the development of cancer drugs for children and adolescents with cancer in the new regulatory environment.
Technical teams together with clinicians across the consortium will define a common data model (CDM) at two levels: First, a smaller set of common data elements (CDE), shared by most or even all data sources, will be used to search for, visualise and query data across all connected partner sites. Secondly, this data set is supplemented by project-specific larger data sets for data integration that address specific research questions. For this second step, common trajectories of scientific questions between the main areas of basic biological research, clinical-biological translational research and clinical studies are defined.

Using the example of bowel cancer, the OnkoFDZ project aims to combine data from seven cancer registries with other health-related medical data such as concomitant diseases, therapies or links to study data. Subsequently, AI methods such as machine learning will be used to capture the use and effectiveness of various therapies and to make the results of the analyses conducted usable for target groups, treatment providers and the public.
Our Software Tools
Going beyond concepts and protoypes, we routinely develop our methods into production-ready software tools, which we – as strong advocates of free open-source software (FOSS) – publish under FOSS licenses on GitHub et al.

Since 2013, bridgeheads have been used as an operating system to ensure secure data work that is both heterogeneous and distributed.
Open source: https://github.com/samply/bridgehead
This technology enables:
- The research and analysis of over 600,000 patients and over 280,000 biosamples
- Linking research data with data from routine clinical care in university medicine
- Improve the availability of molecular therapies
- Identification of similar patients based on molecular markers
- Multinational collaboration between study networks
- Improved accessibility of biospecimens and related clinical data
- Linking of patient records across institutions without sharing sensitive data
- Harmonization of data protection regulations and research needs
- Semantically interoperable data exchange in oncology

The Mainzelliste is a web-based pseudonymization service and was developed as a successor to the PID generator of TMF e.V.
Open Source: https://bitbucket.org/medicalinformatics/mainzelliste
It allows the generation of personal identifiers (PID) from identifying attributes (IDAT), even with changing quality of identifying data thanks to record linkage functionality. Its functions are provided via a REST interface, which enables particularly flexible integration by other software.
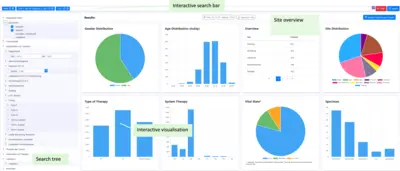
Samply.Lens is a powerful web application specifically designed for the efficient and flexible exploration of federated data.
Open Source: https://github.com/samply/lens
With a focus on performance, interoperability and ease of use, Samply.Lens offers innovative features that improve data discoverability for researchers and enable lightweight analysis and superficial data exploration.
Samply.Lens is already being used successfully in various projects:
- DKTK
- DKTK Joint Funding EXLIQUID
- HiGHMED Use-Case Oncology
- BBMRI
- EUCAIM

TransFAIR is used for data integration in medical facilities and facilitates the ETL process: extraction from source systems, transformation into target schemas and loading into the target system.
Open Source: https://github.com/samply/transFAIR
TransFAIR is especially designed to minimize the effort of data integration for sites that are connected to multiple networks. By supporting new dataset/mapping definitions, these can be easily extended, speeding up the introduction of new functions and dataset extensions. Data quality is improved as errors in the TransFAIR mappings can be corrected centrally.
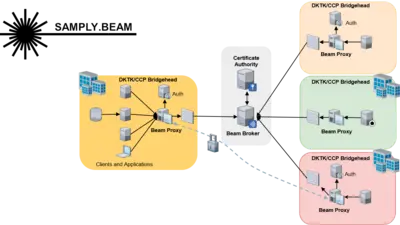
Samply.Beam is designed for efficient and secure network communication in highly restrictive network environments. As a distributed task broker system, it enables the most commonly used communication patterns in virtually all networks, especially those using restrictive firewall rules and exotic proxy servers.
Open Source: https://github.com/samply/beam
Written in Rust, it pursues performance, robustness and security as primary design goals, providing end-to-end encryption and signatures as well as optimized certificate management based on an easy-to-use REST API. Unlike previous middlewares, Beam is better suited for restrictive network settings and can handle the high-bandwidth, low-latency communication required by many SMPC frameworks.
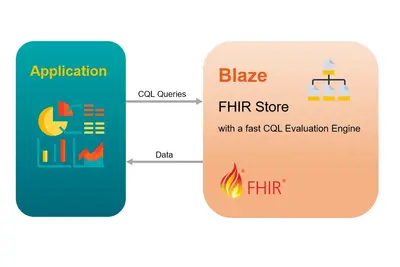
Blaze is a specialized database server for the management and analysis of medical data. It implements the HL7 FHIR specification, an internationally recognized and widely used standard for the exchange and storage of medical data.
Open Source: https://github.com/samply/blaze
Blaze "speaks" the HL7 FHIR language so that it can store and retrieve data structured according to this standard. In addition to FHIR searches, Blaze also supports queries using the Clinical Quality Language (CQL). CQL is a domain-specific language standardized by HL7 that is used to express clinical decision logic and quality measures in a human-readable, computerized manner.
A special feature of Blaze Server is its specific focus on medical applications. It is worth noting that Blaze does not have a user interface, but operates as a background program.
oBDS2FHIR enables seamless data integration, interoperability and scalability for clinical research.
The EpiSelector is a web-based application that supports the selection of comparison groups through matching. The EpiSelector is designed for medical researchers with varying levels of matching expertise. It enables a transparent and reproducible selection of comparison groups and provides step-by-step guidance and recommendations throughout the matching process.
The EpiSelector can be used in combination with other (data preprocessing) components and will be available as a prototype in a Docker container in the near future:
OpenSource: https://github.com/samply/EpiSelector
Team
In our team, we develop and operate national IT infrastructures for the secure exchange of patient data, for example between the university hospitals in the German Consortium for Translational Cancer Research, biomaterial banks of the German Biobank Alliance or the National Network Genomic Medicine Lung Cancer. As a bridging department between the DKFZ and the University Medical Centre Mannheim, we support excellent medical and scientific staff in the transfer of novel cancer therapies into practical application.
-

Prof. Dr. Martin Lablans
Head of Department
-

Nabe Al Hasnawi
-

Moanes Ben Amor
-

Torben Brenner
-

Dr. David Croft
-

Pierre Delpy
-
Leandro Ariel Doctors Lopez
-
Adrian Estevez
-

Claudia Funke
Scientific Project Coordination/Assistance
-

Dennis Grimm
-

Lena Grimm
Assistance
-

Defne Halici
-
Mats Johansen
-

David Juarez Guajardo-Fajardo
-
Martin Jurk
-

Jori Kern
-
Paola Klein
-

Enola Knezevic
-

Thomas Köhler
-
Denis Köther
-

Dr. Eva Krieghoff-Henning
-

Dr. Tobias Kussel
-

Mohamed Lambarki
-
Thewind Mom
-
Valerie Sauer
-

Dr. Esther Schmidt
Deputy Head of Department
-
Tim Schumacher
-

Emil Daniel Simes
-

Jan Skiba
-
Patrick Skowronek
-

Dr. Monique Stenzel
-

Galina Tremper
-
Manoj Waikar
Publications
We would like to thank our sponsors:

Get in touch with us


