Theoretical Systems Biology
- Functional and Structural Genomics

Prof. Dr. Thomas Höfer
We investigate how cancer originates in developing or renewing tissues. To this end, we devise predictive computational models of somatic evolution, stem cell dynamics and immune responses.
Image: Computational reconstruction of neuroblastoma evolution from genome sequencing data (schematic redrawn from G. Caravagna, Mathematical modeling of neuroblastoma associates evolutionary patterns with outcomes, Nat. Genet. 2023)., © dkfz.de
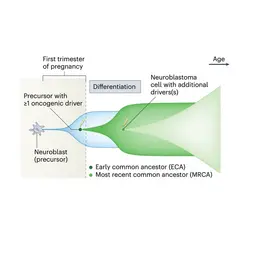
Image: Computational reconstruction of neuroblastoma evolution from genome sequencing data (schematic redrawn from G. Caravagna, Mathematical modeling of neuroblastoma associates evolutionary patterns with outcomes, Nat. Genet. 2023)., © dkfz.de
Our Research

Present research priorities
Given that cancer-associated mutations occur continuously in renewing tissues, such as blood, these tissues are likely to have protective mechanisms in place that limit the selection of cancer drivers and malignant transformation. To understand the nature of this protection, we study the dynamics of non-malignant, steady-state and stress, hematopoiesis in mice and humans, for both cell clones (Busch et al., 2015; Pei et al., 2017) and the underlying molecular regulation. Moreover, using mouse models of sporadic leukemia and human blood and bone marrow samples, we investigate how perturbations in normal stem and progenitor cell dynamics instigate the development of acute leukemia. Methods to study somatic evolution in humans, using population genetics of somatic mutations, carry over to solid tissues, allowing us to study the evolution of cancer in the developing and adult nervous systems (Körber et al., 2023). Our research on clonal dynamics in somatic evolution was initiated by research on T cell responses at single-clone resolution. Here, we devised a mathematical inference framework predicting from experimental data that memory T cells begin to develop early during an acute immune response, from a stem-like T cell subset (Buchholz et al. 2013). This view has now gained broad acceptance, and we continue to dissect the clonal dynamics of adaptive immune responses and their regulation. All our projects involve close collaboration with experimentalists and theoreticians.
Current developments
The computational reconstruction of somatic evolution from genome sequencing data allows us the timing of key genetic events in oncogenesis. In collaboration with researchers at the Hopp Children’s Cancer Center, we are currently exploring the use of these methods for early risk prediction for pediatric cancers. Furthermore, we are developing robust methods to detect clonal selection in normal tissues.
Methods and technologies
We make use of a broad array of methods for the mathematical modeling of clonal dynamics and somatic evolution and for confronting these models with experimental data, using Bayesian inference and artificial intelligence. For in situ barcoding studies in mice, we developed a suite of computational tools, tailored to the Polylox and PolyloxExpress systems (Pei et al., 2017).
Goals and societal relevance
We expect that understanding the mechanisms that allow pre-malignant clones to grow in normal tissues and eventually develop into full-blown malignant tumors will allow us to develop new methods for the earlier detection of cancer, in turn enabling earlier and more effective treatment.
Projects
Stem cell dynamics in development and homeostasis
The rates of stem and progenitor cell differentiation, as well as their regulation in development and homeostasis, cannot be measured directly. We are developing computational methods to infer them from barcoding data in mice or somatic variant statistics in humans. Our focus has been on the hematopoietic system, both on definitive hematopoiesis and the development of tissue macrophages.
Somatic evolution
We are developing methods to infer the somatic evolution of an individual tissue. We are applying these to reconstruct the evolutionary dynamics of cancer in a patient-specific manner and, more recently, have turned to studying mutation and selection dynamics in normal or pre-malignant tissues.
Molecular regulation of hematopoietic stem cell decisions
In close interplay with Project 1 (Stem cell dynamics), we are learning gene-regulatory networks that govern the modes and rates of hematopoietic stem and progenitor cell differentiation and proliferation.
Clonal dynamics of immune responses
T cell immune responses are maintained by small subsets of stem-cell-like T cells. In collaborative projects, we are probing the molecular regulation of stem-cell and effector fate decisions. We are also studying the clonal dynamics of NK cell responses.
Team
-

Prof. Dr. Thomas Höfer
-

Dr. Nils Benjamin Becker
-

Dr. Sarah Benedetto
-
Diana Best
-

Dr. Patrick Binder
-

Nina Claudino
-

Franziska Esau
-
Alessandro Greco
-
Dr. Matthias Günther
-
Philipp Höflich
-
Tatsuro Ikeda
-
Dr. Nikolaus Kepper
-
Emma Kray
-

Maurice Langhinrichs
-
Odette Luber
-

Jonas Metz
-

Tamar Nizharadze
-

Srishti Patil
-
Dr. Giada Sandrini
-
Aisha Schuhegger
-

Dr. Lea Weber
-
Meike Witschel
Selected Publications
Körber V et al.
Körber V et al.
Pei W, Feyerabend TB, et al.
Busch K et al.
Buchholz VR, Flossdorf M, et al.
Get in touch with us

