Microbiome and Cancer
- Immunology, Infection and Cancer

Dr. Eran Elinav
At the Division of Microbiome and Cancer, we study the interactions between the mammalian host, the immune system, and the microbiota.

Our Research
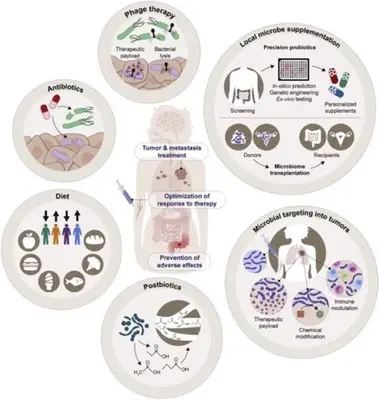
Humans have co-evolved with complex consortium of viruses, bacteria, fungi, and parasites that colonize mucosal surfaces, particularly the gastrointestinal tract, collectively referred to as the microbiota. In mammals, the microbiota can influence a number of physiologic processes, including development, metabolism, and immunological functions. Furthermore, changes in the composition of the microbiota have been associated with an increased susceptibility to a number of diseases, including obesity, asthma, and inflammatory bowel disease. Furthermore, numerous studies have demonstrated a correlation between microorganisms and cancer. Altered local microbiota compositions have been observed in various types of cancer, and specific pathogens, including Helicobacter pylori, Fusobacterium nucleatum, pks+ Escherichia coli, Epstein–Barr virus, and human papilloma viruses, have been linked to carcinogenesis. Additionally, the intestinal microbiota continues to be linked to the efficacy and toxicity of immunotherapies, including immune checkpoint inhibitors, Chimeric Antigen Receptor (CAR) T cell therapy, and allogeneic hematopoietic stem cell transplantation.
At the Division of Microbiome and Cancer, we use both well-established and cutting-edge wet-lab techniques, a state-of-the-art germ-free mouse facility, and novel bioinformatics tools to uncover the relationships between the mammalian host, immune system, and the microbiome. We are elucidating the roles of these factors in carcinogenesis and immunotherapy efficacy while discovering predictive biomarkers for cancers and treatment outcome.
Using translational research approaches we aim to model microbiota perturbations associated with cancer (including breast, brain, colorectal, and pancreatic cancer) in pre-clinical mouse models to understand the mechanisms of microbiota-host cross-talk in these disorders. We investigate how diet or antibiotic-induced microbial dysbiosis affects carcinogenesis in mouse models and study the role of selected microbial isolates, communities of microbes, and microbiota-derived metabolites to understand the molecular mechanisms. We use a variety of well-established and novel bioinformatics tools and software to explore and integrate the wealth of nucleic acid, protein, and metabolite data generated from wet-lab experiments or clinical samples. Advanced computational strategies to integrate metagenomic, transcriptomic, and proteomic data are essential to fully understand and the complexity of host-microbiome interactions in the context of cancer.
Projects
The human microbiome constitutes a complex multikingdom community that symbiotically interacts with the host across multiple body sites. Host-microbiome interactions impact multiple physiological processes and a variety of multifactorial disease conditions. In the past decade, microbiome communities and certain microbial species have been suggested to influence the development, progression, metastasis formation, and treatment response of multiple cancer types. We study the cross-talk between cancer cells, immune cells, the microbiota, metabolism, and other environmental or clinical factors. We hope that discovering new connections between these players involved in the initiation and progression of cancer might lead to the development of new diagnostic and therapeutic approaches.
Our current pre-clinical and clinical research focus includes pancreatic cancer, glioblastoma, and colorectal cancer. We use advanced computational strategies, such as machine learning algorithms run on a high-performance computing cluster, to study microbial communities and the complexity of host-microbiome interactions in the context of cancer.
Team
18 Employees
-

Dr. Eran Elinav
-
Georgios Aindelis
-

Sahana Asokan
PhD Student
-

Dinara Burnasheva
Administrative assistant
-

Laetitia Bury
-

Demet Celik
-

Dr. Arunraj Dhamodaran
-
Louis Dutriat
-

Dr. Rogier Aäron Gaiser
-
Emma Koch
-

Dr. Ignacio Agustin Mastandrea Trias
-
Dr. Gayatree Mohapatra
-

Serena Onwuka
PhD Student
-

Stamatia Papagiannarou
-

Sabine Schmidt
-

Barbara Verhaar
-

Kerstin Wickenheisser
-

Dr. Eli Zamir
Selected Publications
Nobs, S. P.*, Kolodziejczyk, A. A.*, Adler, L., Horesh, N., Botscharnikow, C., Herzog, E., Mohapatra, G., Hejndorf, S., Hodgetts, R.-J., Spivak, I., Schorr, L., Fluhr, L., Kviatcovsky, D., Zacharia, A., Njuki, S., Barasch, D., Stettner, N., Dori-Bachash, M., Harmelin, A., Brandis, A., Mehlman, T., Erez, A., He, Y., Ferrini, S., Puschhof, J., Shapiro, H., Kopf, M., Moussaieff, A., Abdeen, S. K. & Elinav, E.
Siracusa, F., Schaltenberg, N., Kumar, Y., Lesker, T. R., Steglich, B., Liwinski, T., Cortesi, F., Frommann, L., Diercks, B.-P., Bönisch, F., Fischer, A. W., Scognamiglio, P., Pauly, M. J., Casar, C., Cohen, Y., Pelczar, P., Agalioti, T., Delfs, F., Worthmann, A., Wahib, R., Jagemann, B., Mittrücker, H.-W., Kretz, O., Guse, A. H., Izbicki, J. R., Lassen, K. G., Strowig, T., Schweizer, M., Villablanca, E. J., Elinav, E., Huber, S., Heeren, J. & Gagliani, N.
Dougherty, M. W., Valdés-Mas, R., Wernke, K. M., Gharaibeh, R. Z., Yang, Y., Brant, J. O., Riva, A., Muehlbauer, M., Elinav, E., Puschhof, J., Herzon, S. B. & Jobin, C.
Zheng, D., Mohapatra, G., Kern, L., He, Y., Shmueli, M. D., Valdés-Mas, R., Kolodziejczyk, A. A., Próchnicki, T., Vasconcelos, M. B., Schorr, L., Hertel, F., Lee, Y. S., Rufino, M. C., Ceddaha, E., Shimshy, S., Hodgetts, R. J., Dori-Bachash, M., Kleimeyer, C., Goldenberg, K., Heinemann, M., Stettner, N., Harmelin, A., Shapiro, H., Puschhof, J., Chen, M., Flavell, R. A., Latz, E., Merbl, Y., Abdeen, S. K. & Elinav, E.
Stein-Thoeringer, C. K., Saini, N. Y., Zamir, E., Blumenberg, V., Schubert, M.-L., Mor, U., Fante, M. A., Schmidt, S., Hayase, E., Hayase, T., Rohrbach, R., Chang, C.-C., McDaniel, L., Flores, I., Gaiser, R., Edinger, M., Wolff, D., Heidenreich, M., Strati, P., Nair, R., Chihara, D., Fayad, L. E., Ahmed, S., Iyer, S. P., Steiner, R. E., Jain, P., Nastoupil, L. J., Westin, J., Arora, R., Wang, M. L., Turner, J., Menges, M., Hidalgo-Vargas, M., Reid, K., Dreger, P., Schmitt, A., Müller-Tidow, C., Locke, F. L., Davila, M. L., Champlin, R. E., Flowers, C. R., Shpall, E. J., Poeck, H., Neelapu, S. S., Schmitt, M., Subklewe, M., Jain, M. D., Jenq, R. R. & Elinav, E.
Get in touch with us

