Software
Bridgehead

Since 2013, bridgeheads have been used as an operating system to ensure secure data work that is both heterogeneous and distributed.
Open source: https://github.com/samply/bridgehead
This technology enables:
- The research and analysis of over 600,000 patients and over 280,000 biosamples
- Linking research data with data from routine clinical care in university medicine
- Improve the availability of molecular therapies
- Identification of similar patients based on molecular markers
- Multinational collaboration between study networks
- Improved accessibility of biospecimens and related clinical data
- Linking of patient records across institutions without sharing sensitive data
- Harmonization of data protection regulations and research needs
- Semantically interoperable data exchange in oncology
Federated Data Analysis with the CCP Platform
The CCP platform offers an innovative solution for cross-site federated data analysis with DataSHIELD. The integration into the CCP platform successfully solves the previous limitations of DataSHIELD. This simplifies the handling of installation, updates and network configurations and provides users with a seamless experience. In addition, RStudio is integrated, providing researchers with a familiar and powerful development environment. This technology allows researchers to analyse data across different locations without having to physically move the data. Instead, the algorithms travel to the data. Only the requested and approved data cohorts are analysed, ensuring the highest standards of data security and privacy. These standards are guaranteed by the integration into the Beam infrastructure from the bridgehead.
Mainzelliste

The Mainzelliste is a web-based pseudonymization service and was developed as a successor to the PID generator of TMF e.V.
Open Source: https://bitbucket.org/medicalinformatics/mainzelliste
It allows the generation of personal identifiers (PID) from identifying attributes (IDAT), even with changing quality of identifying data thanks to record linkage functionality. Its functions are provided via a REST interface, which enables particularly flexible integration by other software.
Samply.Lens
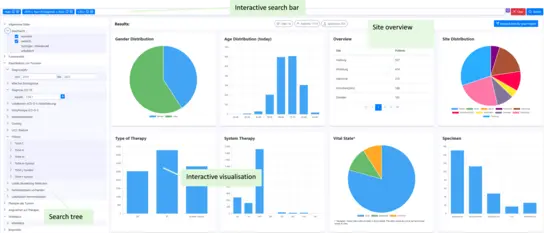
Samply.Lens is a powerful web application specifically designed for the efficient and flexible exploration of federated data.
Open Source: https://github.com/samply/lens
With a focus on performance, interoperability and ease of use, Samply.Lens offers innovative features that improve data discoverability for researchers and enable lightweight analysis and superficial data exploration.
Samply.Lens is already being used successfully in various projects:
- DKTK
- DKTK Joint Funding EXLIQUID
- HiGHMED Use-Case Oncology
- BBMRI
- EUCAIM
Samply.TransFair

TransFAIR is used for data integration in medical facilities and facilitates the ETL process: extractionfrom source systems, transformationinto target schemas and loadinginto the target system.
Open Source: https://github.com/samply/transFAIR
TransFAIR is especially designed to minimize the effort of data integration for sites that are connected to multiple networks. By supporting new dataset/mapping definitions, these can be easily extended, speeding up the introduction of new functions and dataset extensions. Data quality is improved as errors in the TransFAIR mappings can be corrected centrally.
Samply.Beam
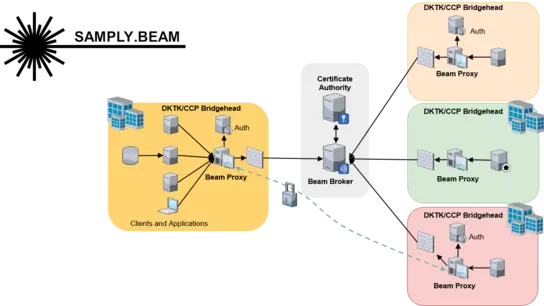
Samply.Beam is designed for efficient and secure network communication in highly restrictive network environments. As a distributed task broker system, it enables the most commonly used communication patterns in virtually all networks, especially those using restrictive firewall rules and exotic proxy servers.
Open Source: https://github.com/samply/beam
Written in Rust, it pursues performance, robustness and security as primary design goals, providing end-to-end encryption and signatures as well as optimized certificate management based on an easy-to-use REST API. Unlike previous middlewares, Beam is better suited for restrictive network settings and can handle the high-bandwidth, low-latency communication required by many SMPC frameworks.
Samply.Blaze
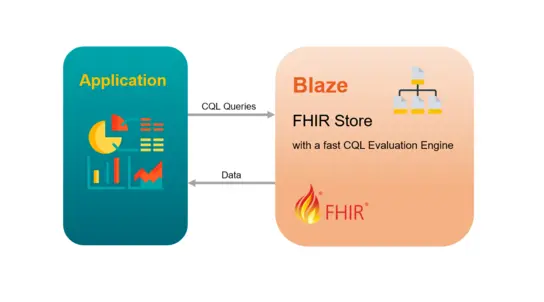
Blaze is a specialized database server for the management and analysis of medical data. It implements the HL7 FHIR specification, an internationally recognized and widely used standard for the exchange and storage of medical data.
Open Source: https://github.com/samply/blaze
Blaze "speaks" the HL7 FHIR language so that it can store and query data structured according to this standard. In addition to FHIR searches, Blaze also supports queries using the Clinical Quality Language (CQL). CQL is a domain-specific language standardized by HL7 that is used to express clinical decision logic and quality measures in a human-readable, computerized manner.
A special feature of Blaze Server is its specific focus on medical applications. It is worth noting that Blaze does not have a user interface, but operates as a background program.
Samply.oBDS2FHIR
oBDS2FHIR is a specialized ETL solution(Extract, Transform, Load) for the automated conversion of the Oncological Basic Data Set (oBDS) into the FHIR format. The tool enables German university hospitals to efficiently integrate their oncology registry data into research networks without having to develop their own ETL processes.
Open Source: https://github.com/samply/obds2fhir
Research data integration goes beyond a purely technical conversion (see Samply.TransFair). On the one hand, a research-oriented model is derived from the tumor documentation. On the other hand, the data is harmonized and its content coordinated, as the oBDS is defined as a standard but is applied inconsistently and interpreted differently in practice. This is done taking into account the real differences in the tumor documentation of German university hospitals in order to create a comparable database.
EpiSelector
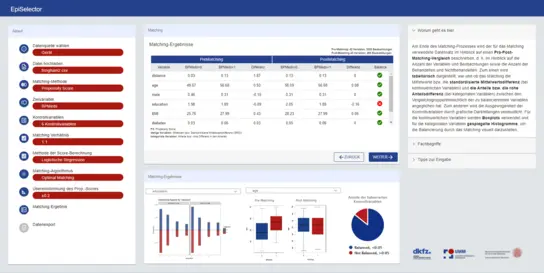
The EpiSelector is a web-based application that supports the selection of comparison groups through matching. The EpiSelector is designed for medical researchers with varying levels of matching expertise. It enables a transparent and reproducible selection of comparison groups and provides step-by-step guidance and recommendations throughout the matching process.
The EpiSelector can be used in combination with other (data preprocessing) components and will be available as a prototype in a Docker container in the near future:
OpenSource: https://github.com/samply/EpiSelector