Clinical Cooperation Unit Neurooncology
Research of the clinical cooperation unit (CCU) neurooncology has the overarching goal to more comprehensively understand the biologic mechanisms underlying glioblastoma and primary CNS lymphoma and their respective treatment resistance. We strive for the identification and validation of diagnostic, prognostic and predictive biomarkers for guiding clinical decision-making and ultimately develop new points/molecules for therapeutic intervention that are to be tested in follow-up clinical trials.

Research
The current and future focus of the research in the Neurooncology Clinical Cooperation Unit (CCU) is based on central research questions derived from Heidelberg University Hospital neurooncology program, with the clear aim of translating the pre-clinical observations back into the clinic in a bench-to-bedside approach. Our CCU Neurooncology is strongly linked to the CCU Neuroimmunology and Brain Tumor Immunology (D170), CCU Neuropathology (B300) and has vital collaborations to the Department of Hematology, Oncology, and Rheumatology as well as to the experimental imaging departments at both the Head Clinic and the DKFZ. Moreover, close collaborations exist within the UNITE glioblastoma consortium (https://www.unite-glioblastoma.de/). Integration into a national as well as international network additionally strengthens and is of utmost importance for the rapid transfer of research results to the clinic.
Projects
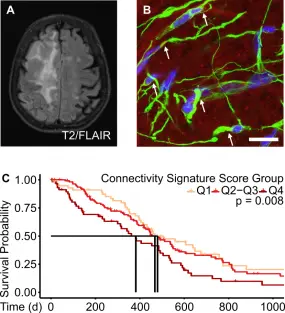
With the exciting discovery of multicellular glioblastoma networks a new concept of growth and resistance for brain tumors has been established. Aiming to decipher the molecular underpinnings of these networks we established a transcriptomic signature for connected glioblastoma cells through single cell and bulk RNA sequencing of in vitro and in vivo models as well as patient tissue from clinical studies. We identified chitinase-3-like 1 (CHI3L1), a molecular marker with functional relevance for tumor cell networks and potential therapeutic applicability, in glioblastoma and validated its significance. These discoveries have pathed the way for further follow-up projects on the exact mechanism of action of CHI3L1 and the relevance of other putative tumor network relevant genes.
This discovery arm aims to unravel the molecular mechanisms of several targeted therapies such as Temsirolimus, an mTOR inhibitor, and Atezolizumab, an anti-PD-L1 antibody, but also to define pathways involved in acquired resistance, such as TP53 signaling for alkylating therapy. We also aim to understand the interaction between radiation and the molecularly defined treatments by analyzing patient tissue with omics technologies in the scope of the NCT Neuro Master Match (N2M2) umbrella trial for newly diagnosed MGMT unmethylated glioblastoma and other larger randomized trials coordinated by the clinical neurooncology unit.
This consists of a multiomics integrative approach including cytology, proteomics and ctDNA sequencing from cerebrospinal fluid (CSF). Harnessing proteomics has allowed grouping of glioblastoma patients into prognostically different clusters and identification of CSF biomarkers that are unique for glioblastoma compared to other brain malignancies. A cfDNA sequencing platform analyzing a panel of more than 200 brain tumor relevant genes was established and is currently capable of detecting mutations and copy number profiles in glioblastoma patients, potentially allowing a diagnosis without biopsy in difficult to biopsy conditions or tracing tumor evolution.
Molecular fingerprints and phenotypic traits of glioblastoma stem cells.
Immune checkpoint inhibitors, tumor vaccines as well as CAR-T cell approaches are current object of research to address glioblastoma-enriched or specific targets in both the primary and recurrent setting. Specifically, we investigate intervention therapies for PD-L1, IDH1 R132H, and additional drug targets upregulated after standard of care therapy.
In previous studies we have applied unbiased genetic approaches (Whole Exome/Genome Sequencing, 850k analysis, among others) to PCNSL specimen in order to define molecular hallmarks of CNS lymphomagenesis. Yet many detected alterations remain incompletely understood. Findings are therefore modeled in lymphoma cell lines in vitro and in vivo using xenograft models. The latter allows to uncover the phenotypical and molecular consequences of various genetic alterations. In vitro drug screens and CAR-T cell engineering allow to guide novel targeted therapies, that are initially evaluated in a preclinical mouse model, with an aim for translation into early clinical trials.
Team
-

Prof. Dr. med. Wolfgang Wick
Head of the Division
-

Dr. Tobias Kessler
Scientist
-

Robin Wagener
Scientist
-

Dr. Leon Kaulen
Scientist
-

Dirk Hoffmann
Scientist
-

Dr. Uwe Warnken
Scientist
-

Alexandros Kourtesakis
Scientist
-

Hiu Nam Hannah Chow
PhD Student
-

Melissa Hahn
PhD Student
-

Hannah Rohdjeß
PhD Student
-
Andy Schacht
PhD Student
-

Sebastian Schulz
MD Student
-

Normann Mußnig
MD Student
-

Tim Windheim
MD Student
-

Gina Marie Cebulla
MD Student
-
Florian Iser
-

Disha Biswas
Master's Student
-

Sonja Pusch
CTA
-

Petra Rübmann
MTA
-

Denise Reibold
BTA (on parental leave)
-
Lena Ehret-Maßholder
BTA
-
Melanie Kuhse
BTA
-

Zuoyi Xu
Research Assistant
-

Lisa-Maria Glatzel
Research Assistant
Publications
Wolfgang Wick, Lisa-Marie Lanz, Antje Wick, Inga Harting, Susan Dettmer, Abigail K. Suwala, Ralf Ketter, Ghazaleh Tabatabai, Corinna Seliger, Martin Glas, Michael C. Burger, Marco Timmer, Florian A. Ringel, Iris Mildenberger, Walter J. Schulz-Schaeffer, Frank Winkler, Laila König, Christel Herold-Mende, Andreas Eisenmenger, Stefan M. Pfister, Mirjam Renovanz, Martin Bendszus, Felix Sahm, Michael Platten, Tobias Kessler
Kats, I. ; Simovic-Lorenz, M. ; Schreiber, H. ; Sant, P. ; Mallm, J.-P. ; Körber, V. ; Li, A. ; Velmurugan, P. ; Heuer, S. ; Kües, L. ; Devens, F. ; Sill, M. ; Jugold, M. ; Moustafa, M. ; Abdollahi, A. ; Winkler, F. ; Korshunov, A. ; Pfister, S. ; Stegle, O. ; Ernst, A.
Deng, M. Y. ; Rauh, S. ; Anil, G. ; Lischalk, J. W. ; Hahnemann, L. ; Eichkorn, T. ; Hörner-Rieber, J. ; Paul, A. ; Sandrini, E. ; Hoegen-Sassmannshausen, P. ; Held, T. ; Regnery, S. ; Bauer, L. ; Sahm, F. ; von Deimling, A. ; Wick, A. ; Wick, W. ; Jungk, C. ; Krieg, S. M. ; Herfarth, K. ; Debus, J. ; König, L.
Alhalabi, O. ; Göttmann, M. ; Gold, M. P. ; Schlue, S. ; Hielscher, T. ; Iskar, M. ; Kessler, T. ; Hai, L. ; Lokumcu, T. ; Cousins, C. C. ; Herold-Mende, C. ; Heßling, B. ; Horschitz, S. ; Jabali, A. ; Koch, P. ; Baumgartner, U. ; Day, B. W. ; Wick, W. ; Sahm, F. ; Krieg, S. M. ; Fraenkel, E. ; Phillips, E. ; Goidts, V.
Schubert, M. C. ; Soyka, S. J. ; Tamimi, A. ; Maus, E. ; Schroers, J. ; Wißmann, N. ; Reyhan, E. ; Tetzlaff, S. K. ; Yang, Y. ; Denninger, R. ; Peretzke, R. ; Beretta, C. ; Drumm, M. ; Heuer, A. ; Buchert, V. ; Steffens, A. ; Walshon, J. ; McCortney, K. ; Heiland, S. ; Bendszus, M. ; Neher, P. ; Golebiewska, A. ; Wick, W. ; Winkler, F. ; Breckwoldt, M. O. ; Kreshuk, A. ; Kuner, T. ; Horbinski, C. ; Kurz, F. T. ; Prevedel, R. ; Venkataramani, V.
Sievers, P. ; Bielle, F. ; Göbel, K. ; Schrimpf, D. ; Nichelli, L. ; Mathon, B. ; Appay, R. ; Boldt, H. B. ; Dohmen, H. ; Selignow, C. ; Acker, T. ; Vicha, A. ; Martinetto, H. ; Schweizer, L. ; Schüller, U. ; Brandner, S. ; Wesseling, P. ; Schmid, S. ; Capper, D. ; Abdullaev, Z. ; Aldape, K. ; Korshunov, A. ; Krieg, S. M. ; Wick, W. ; Pfister, S. M. ; von Deimling, A. ; Reuss, D. E. ; Jones, D. ; Sahm, F.
Streibel, Y. ; Breckwoldt, M. ; Hunger, J. ; Pan, C. ; Fischer, M. ; Turco, V. ; Boztepe, B. ; Fels-Palesandro, H. ; Scheck, J. G. ; Sturm, V. ; Karimian-Jazi, K. ; Agardy, D. A. ; Annio, G. ; Mustapha, R. ; Soni, S. S. ; Alasa, A. ; Weidenfeld, I. ; Rodell, C. B. ; Wick, W. ; Heiland, S. ; Winkler, F. ; Platten, M. ; Bendszus, M. ; Sinkus, R. ; Schregel, K.
Ellingson, B. M. ; Sanvito, F. ; Cloughesy, T. F. ; Huang, R. Y. ; Villanueva-Meyer, J. E. ; Pope, W. B. ; Barboriak, D. P. ; Shankar, L. K. ; Smits, M. ; Kaufmann, T. J. ; Boxerman, J. L. ; Weller, M. ; Galanis, E. ; Groot, J. d. ; Gilbert, M. R. ; Lassman, A. B. ; Shiroishi, M. S. ; Nabavizadeh, A. ; Mehta, M. ; Stupp, R. ; Wick, W. ; Reardon, D. A. ; Vogelbaum, M. A. ; van den Bent, M. ; Chang, S. M. ; Wen, P. Y.
Hegi, M. E. ; Oppong, F. B. ; Perry, J. R. ; Wick, W. ; Henriksson, R. ; Laperriere, N. J. ; Gorlia, T. ; Malmström, A. ; Weller, M. ; NCBTSG ; NOA ; CCTG ; EORTC-BTG
Hu, Y. ; Hruscha, A. ; Pan, C. ; Schifferer, M. ; Schmidt, M. K. ; Nuscher, B. ; Giera, M. ; Kostidis, S. ; Burhan, Ö. ; van Bebber, F. ; Edbauer, D. ; Arzberger, T. ; Haass, C. ; Schmid, B.
Park, Y. W. ; Choi, K. S. ; Foltyn-Dumitru, M. ; Brugnara, G. ; Banan, R. ; Kim, S. ; Han, K. ; Park, J. E. ; Kessler, T. ; Bendszus, M. ; Krieg, S. ; Wick, W. ; Sahm, F. ; Choi, S. H. ; Kim, H. S. ; Chang, J. H. ; Kim, S. H. ; Wongsawaeng, D. ; Pollock, J. M. ; Lee, S.-K. ; Barajas, R. F. ; Vollmuth, P. ; Ahn, S. S.
Roesler, J. ; Spitzer, D. ; Jia, X. ; Aasen, S. N. ; Sommer, K. ; Roller, B. ; Olshausen, N. ; Hebach, N. R. ; Albinger, N. ; Ullrich, E. ; Zhu, L. ; Wang, F. ; Macas, J. ; Forster, M.-T. ; Steinbach, J. P. ; Sevenich, L. ; Devraj, K. ; Thorsen, F. ; Karreman, M. A. ; Plate, K. H. ; Reiss, Y. ; Harter, P.
Deng, M. Y. ; da Silva, A. S. ; Göller, P. ; König, L. ; Schäfer, H. ; Maire, C. ; Lentz-Hommertgen, A. ; Held, T. ; Regnery, S. ; Eichkorn, T. ; Stritzke, F. ; Bauer, L. ; Schnell, D. ; Herfarth, K. ; von Deimling, A. ; Krieg, S. ; Wick, A. ; Wick, W. ; Grosu, A. ; Debus, J. ; Sahm, F. ; Ricklefs, F.
Amit, M. ; Anastasaki, C. ; Dantzer, R. ; Demir, I. E. ; Deneen, B. ; Dixon, K. O. ; Egeblad, M. ; Gibson, E. M. ; Hervey-Jumper, S. L. ; Hondermarck, H. ; Magnon, C. ; Monje, M. ; Na'ara, S. ; Pan, Y. ; Repasky, E. A. ; Scheff, N. N. ; Sloan, E. K. ; Talbot, S. ; Tracey, K. J. ; Trotman, L. C. ; Valiente, M. ; Van Aelst, L. ; Venkataramani, V. ; Venkatesh, H. S. ; Vermeer, P. D. ; Winkler, F. ; Wong, R. J. ; Gutmann, D. H. ; Borniger, J. C.
Swanton, C. ; Bernard, E. ; Abbosh, C. ; André, F. ; Auwerx, J. ; Balmain, A. ; Bar-Sagi, D. ; Bernards, R. ; Bullman, S. ; DeGregori, J. ; Elliott, C. ; Erez, A. ; Evan, G. ; Febbraio, M. A. ; Hidalgo, A. ; Jamal-Hanjani, M. ; Joyce, J. A. ; Kaiser, M. ; Lamia, K. ; Locasale, J. W. ; Loi, S. ; Malanchi, I. ; Merad, M. ; Musgrave, K. ; Patel, K. J. ; Quezada, S. ; Wargo, J. A. ; Weeraratna, A. ; White, E. ; Winkler, F. ; Wood, J. N. ; Vousden, K. H. ; Hanahan, D.
Rastogi, A. ; Brugnara, G. ; Foltyn-Dumitru, M. ; Mahmutoglu, M. A. ; Preetha, C. J. ; Kobler, E. ; Pflüger, I. ; Schell, M. ; Deike-Hofmann, K. ; Kessler, T. ; van den Bent, M. J. ; Idbaih, A. ; Platten, M. ; Brandes, A. A. ; Nabors, B. ; Stupp, R. ; Bernhardt, D. ; Debus, J. ; Abdollahi, A. ; Gorlia, T. ; Tonn, J.-C. ; Weller, M. ; Maier-Hein, K. ; Radbruch, A. ; Wick, W. ; Bendszus, M. ; Meredig, H. ; Kurz, F. T. ; Vollmuth, P.
Huppertz, M.-C. ; Wilhelm, J. ; Grenier, V. ; Schneider, M. W. ; Falt, T. ; Porzberg, N. ; Hausmann, D. ; Hoffmann, D. C. F. ; Hai, L. ; Tarnawski, M. ; Pino, G. ; Slanchev, K. ; Kolb, I. ; Acuna, C. ; Fenk, L. M. ; Baier, H. ; Hiblot, J. ; Johnsson, K.
Alhalabi, O. T. ; Dao Trong, P. ; Kaes, M. ; Jakobs, M. ; Kessler, T. ; Oehler, H. ; König, L. ; Eichkorn, T. ; Sahm, F. ; Debus, J. ; von Deimling, A. ; Wick, W. ; Wick, A. ; Krieg, S. M. ; Unterberg, A. W. ; Jungk, C.
Hai, L. ; Hoffmann, D. C. F. ; Wagener, R. J. ; Azorin, D. D. ; Hausmann, D. ; Xie, R. ; Huppertz, M.-C. ; Hiblot, J. ; Sievers, P. ; Heuer, S. ; Ito, J. ; Cebulla, G. ; Kourtesakis, A. ; Kaulen, L. D. ; Ratliff, M. ; Mandelbaum, H. ; Jung, E. ; Jabali, A. ; Horschitz, S. ; Ernst, K. ; Reibold, D. ; Warnken, U. ; Venkataramani, V. ; Will, R. ; Suvà, M. L. ; Herold-Mende, C. ; Sahm, F. ; Winkler, F. ; Schlesner, M. ; Wick, W. ; Kessler, T.
Iser, F. ; Hinz, F. ; Hoffmann, D. C. F. ; Grassl, N. ; Güngör, C. ; Meyer, J. ; Dörner, L. ; Hofmann, L. ; Kelbch, V. ; Göbel, K. ; Mahmutoglu, M. A. ; Vollmuth, P. ; Patel, A. J. ; Nguyen, D. ; Kaulen, L. ; Mildenberger, I. ; Sahm, K. ; Maass, K. ; Pajtler, K. ; Shankar, G. M. ; Weiler, M. ; Wildemann, B. ; Winkler, F. ; von Deimling, A. ; Platten, M. ; Wick, W. ; Sahm, F. ; Kessler, T.
Heuer, S. ; Burghaus, I. ; Gose, M. ; Kessler, T. ; Sahm, F. ; Vollmuth, P. ; Venkataramani, V. ; Hoffmann, D. ; Schlesner, M. ; Ratliff, M. ; Hopf, C. ; Herrlinger, U. ; Ricklefs, F. ; Bendszus, M. ; Krieg, S. M. ; Wick, A. ; Wick, W. ; Winkler, F.
Deng, M. Y. ; Maas, S. L. N. ; Hinz, F. ; Karger, C. P. ; Sievers, P. ; Eichkorn, T. ; Meixner, E. ; Hoegen-Sassmannshausen, P. ; Hörner-Rieber, J. ; Lischalk, J. W. ; Seidensaal, K. ; Bernhardt, D. ; Jungk, C. ; Unterberg, A. ; Wick, A. ; Wick, W. ; von Deimling, A. ; Sahm, F. ; Combs, S. ; Herfarth, K. ; Debus, J. ; König, L.
Derby, S. ; Dutton, L. ; Strathdee, K. E. ; Stevenson, K. ; Koessinger, A. ; Jackson, M. ; Tian, Y. ; Yu, W. ; Mclay, K. ; Misquitta, J. ; Alsharif, S. ; Clarke, C. J. ; Gilmour, L. ; Thomason, P. ; McGhee, E. ; McGarrity-Cottrell, C. L. ; Vanderlinden, A. ; Collis, S. J. ; Rominyi, O. ; Lemgruber, L. ; Solecki, G. ; Olson, M. ; Winkler, F. ; Carlin, L. M. ; Heiland, D. H. ; Inman, G. ; Chalmers, A. J. ; Norman, J. C. ; Carruthers, R. ; Birch, J. L.
Foesleitner, O. ; Sturm, V. ; Hayes, J. ; Weiler, M. ; Sam, G. ; Wildemann, B. ; Wick, W. ; Bendszus, M. ; Heiland, S. ; Jäger, L. B.
Kessler, T. ; Sam, G. ; Wick, W. ; Weiler, M.
Krämer, C. ; Kilian, M. ; Chih, Y. C. ; Kourtesakis, A. ; Hoffmann, D. C. ; Boschert, T. ; Koopmann, P. ; Sanghvi, K. ; De Roia, A. ; Jung, S. ; Jähne, K. ; Day, B. ; Shultz, L. D. ; Ratliff, M. ; Harbottle, R. ; Green, E. W. ; Will, R. ; Wick, W. ; Platten, M. ; Bunse, L.
Karimian-Jazi, K. ; Vollherbst, D. F. ; Schwarz, D. ; Fischer, M. ; Schregel, K. ; Bauer, G. ; Kocharyan, A. ; Sturm, V. ; Neuberger, U. ; Jesser, J. ; Herweh, C. ; Ulfert, C. ; Hilgenfeld, T. ; Seker, F. ; Preisner, F. ; Schmitt, N. ; Charlet, T. ; Hamelmann, S. ; Sahm, F. ; Heiland, S. ; Wick, W. ; Ringleb, P. A. ; Schirmer, L. ; Bendszus, M. ; Möhlenbruch, M. A. ; Breckwoldt, M. O.
Get in touch with us

