Cellular Immunology
- Immunology, Infection and Cancer

Prof. Dr. Hans-Reimer Rodewald
Leiter der Arbeitsgruppe/Principal Investigator
The Division of Cellular Immunology investigates cell and organ development in the immune system as well as their immunological functions.
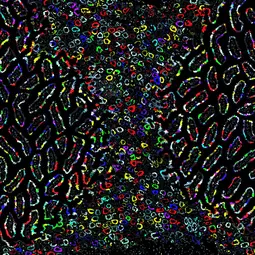
Our Research
The Division of Cellular Immunology investigates the development of cells and organs of the immune system as well as their functions. In this context, we focus on key open questions in physiology and pathology, including the dynamic processes of hematopoietic stem and progenitor cell differentiation under steady state and challenge conditions. Furthermore, we investigate ontogeny and functions of tissue-resident macrophages, innate lymphoid cells, and mast cells. In the context of cancer biology, we have a strong interest in cellular and molecular events underlying the origin of T-ALL, and metastasis of melanoma.
The foundation for our current projects was laid by our previous studies enabling fate-mapping to reveal the physiological output of hematopoietic stem cells (HSCs) in situ (Busch et al. Nature 2015), deciphering the embryonic origin of tissue-resident macrophages (Gomez Perdiguero, Klapproth et al. Nature 2015), and establishing a specifically mast cell-deficient mouse mutant (Feyerabend et al. Immunity 2011). Moreover, our discovery that disruption of cell competition in the thymus predisposes for T-cell acute lymphoblastic leukemia (Martins et al. Nature 2014) set the stage for studies into leukemogenesis.
More recently, we have developed lineage-tracing mouse models that have pushed the resolution of fate-mapping from few colors to several hundred thousand genetically inheritable tags. Starting with “Polylox”, an inducible endogenous DNA barcoding mouse model (Pei, Feyerabend et al. Nature 2017), the system has been advanced towards RNA barcoding (“PolyloxExpress”) to achieve the direct linkage between single cell fates and transcriptomes (Pei, Shang, Wang et al. Cell Stem Cell 2020), and, most recently, epitope barcoding (“Polytope”) for spatial clonal resolution (Postrach et al., bioRxiv 2024).
With these and other experiments, we address in ongoing studies the projects, including those further specified below:
Fundamental properties of unperturbed and perturbed hematopoiesis
High-resolution barcoding
Leukemia development and progression
Mast cell function
Projects
Fundamental properties of unperturbed and perturbed hematopoiesis
Hematopoietic stem cells (HSC) have been widely studied by transplantation into immune- and blood-cell-depleted recipients. To address HSC functions under physiological, i.e. native conditions, we generated a Cre knock-in mouse (Tie2MerCreMer) enabling inducible genetic labeling of HSC during embryonic development and in the adult bone marrow. These experiments showed, unexpectedly, that only in the order of 1% of HSC contributes to hematopoiesis per day. Many HSC contribute infrequently, which is in contrast to earlier conclusions that few HSC contribute strongly. Hence, in situ HSC fate mapping revealed fundamental differences between steady-state maintenance of the hematopoietic system and its re-establishment after transplantation and provided a quantitative framework for studying hematopoiesis in health and disease in situ (Busch et al. Nature 2015).
Our ongoing research resolved the response of stem and progenitor compartments to various challenges, including polymicrobial sepsis, gaining insights into the kinetics and the differentiation processes of perturbed hematopoiesis (Fanti, Busch et al. Cell Stem Cell 2023).
We complement our studies on hematopoiesis at the mechanistic level by investigating the role of epigenetic regulation focusing on the histone deubiquitinase USP22. We found that in the absence of infection or inflammation, blood and immune cell-specific Usp22 deficiency results in the spontaneous, strictly cell-intrinsic activation of systemic emergency hematopoiesis and enhanced anti-bacterial innate immune protection (Dietlein et al. Sci Immunol 2022).
High-resolution barcoding
To decipher hematopoiesis and the immune system at single cell and clonal levels our laboratory has developed Cre-loxP-based DNA and RNA barcoding technologies, termed 'Polylox' and 'PolyloxExpress', respectively (Pei, Feyerabend et al. Nature 2017; Pei, Wang, Rössler et al. Nature Protocols 2019; Pei, Shang, Wang et al. Cell Stem Cell 2020; Fanti, Busch et al. Cell Stem Cell 2023).
Application of these new endogenous barcoding technologies led, among other findings, to the classification of HSC fates into distinct classes, i.e. multilineage, myeloid-erythroid restricted, and differentiation-inactive. The second generation of our barcoding system, 'PolyloxExpress' facilitates simultaneous recovery of barcodes and transcriptomic information at the single cell level. This allowed us to directly link cell fate output with underlying transcriptomic signatures (Pei, Shang, Wang et al. Cell Stem Cell 2020).
We designed ‘Polytope’ to link spatial information and clonal lineage tracing in situ (Postrach et al. bioRxiv 2024). Polytope has 512 distinct color codes, which can be visualized by conventional fluorescence microscopy, providing a comprehensive tool for capturing complex clonal dynamics in situ. For proof of principle, we traced the fate of hundreds of clones across tissues from embryonic development to adulthood.
Overall, our ongoing and future work aims to apply these technologies to further dissect general mechanisms of hematopoietic progenitor cell fate decisions in a multi-omics approach and specifically developmental ontogenies of tissue-resident immune cell subsets, such as macrophages (Frank, Postrach, Langhinrichs et al. bioRxiv 2024), and innate lymphoid cells (Cirovic et al. in preparation) in situ.
Leukemia development and progression
In previous work, we identified cell competition between 'young' bone marrow-derived and 'old' thymus-resident progenitors as a tumor suppressor mechanism for T-cell acute lymphoblastic leukemia (T-ALL) (Martins et al. Nature 2014). Disruption of new influx of progenitors into the thymus leads to spontaneous development of full-blown T-ALL and provides a unique and robust system to unravel cellular and molecular events along the transition from normal to leukemic T cells, closely resembling the human disease (Martins et al. Nature 2014; Ginn et al. Mol Ther Nucleic Acids 2017; Schiroli et al. Sci Transl Med 2017; Gao J et al. Dis Model Mech 2019). As this tumor model is independent of a priori introduced oncogenes, it makes the 'natural emergence' of clones and mutations kinetically accessible and, in general, presents arguably a unique tool to address mechanisms of tumor development and progression.
Along these lines, we apply this system of tumor development in the following contexts:
Investigation of Notch1 driver mutations as rate-limiting step of tumorigenesis
Modeling of stem cell output on the risk of acquiring oncogenic drivers and leukemia progression (in collaboration with the laboratory of Thomas Höfer)
- Deconvolution of metastasis behavior of T-ALL by applying Polylox and PolyloxExpress barcoding and lineage tracing
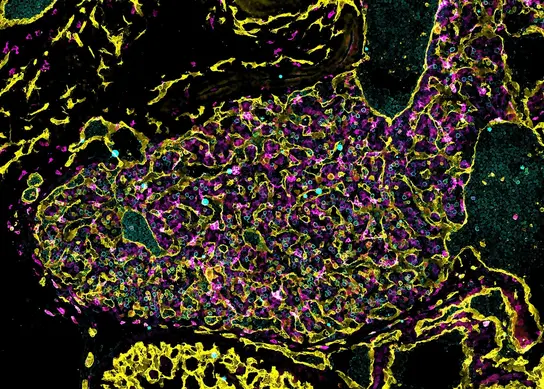
Mast Cell Biology
Mast cells remain highly enigmatic cells of the immune system, and we developed mouse mutants and databases to dissect mast cell biology in health and disease. These include mast cell-deficient mice (Feyerabend et al. Immunity 2011; Rodewald and Feyerabend, Immunity 2012) and proteome of primary human and murine mast cells (Plum et al. Immunity 2020).
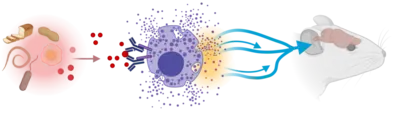
Mast cells promote allergic diseases and are tightly integrated in type 2 immunity. While the immunological underpinnings of allergies are well characterized, the mechanisms of the neurological symptoms of allergies, i.e. itch, cough, emesis, diarrhea, and sneezing, are less well understood. Due to their location at barrier tissues and ability to sense environmental antigens, we investigated the possible roles of mast cells as a link between specific environmental cues and the nervous system. We identified mast cells as crucial mediators in signaling antigen avoidance behavior, thereby preventing or reducing inflammation caused by repeated exposure (Plum, Binzberger et al. Nature 2023). We propose that mast cells rapidly convert these diverse tissue signals into appropriate, organ-specific protective reflexes that can limit inflammation or reduce tissue damage (Plum et al. Immunity 2024).
Team
-
Prof. Dr. Hans-Reimer Rodewald
-
Ulrike Esslinger
Secretary
-
Dr. Thorsten Feyerabend
Senior Scientist
-
Katrin Busch
Postdoc
-
Dr. Nikolaus Dietlein
Postdoc
-
Dr. Thomas Plum
Postdoc
-
Larissa Frank
Ph.D. Student
-
Fuwei Shang
Postdoc
-
Daniel Postrach
Ph.D. Student
-
Robin Thiele
Ph.D. Student
-
Natascha Bäuerle
Ph.D. Student
-
Sven Schäfer
Technical Assistant
-
Günter Küblbeck
Technical Assistant
-
Nicole Maltry
Technical Assistant
-
Celine Beyersdörffer
Technical Assistant
-
Carmen Henrich-Kellner
Technical Assistant
Selected Publications
Plum P, Binzberger R, Thiele R, Shang F, Postrach, Fung C, Fortea M, Stakenborg N, Wang Z, Tappe-Theodor A, Poth T, MacLaren D, Boeckxstaens G, Kuner R, Pitzer C, Monyer H, Xin C, Bonventre J, Tanaka S, Voehringer D, Vanden Berghe P, Strid J, Feyerabend T, Rodewald HR
Fanti AK, Busch K, Greco A, Wang X, Cirovic B, Shang F, Nizharadze T, Frank L, Barile M, Feyerabend T, Höfer T, Rodewald HR
Dietlein N, Wang X, Metz J, Disson O, Shang F, Beyersdörffer C, Rodríguez Correa E, Lipka D B, Begus-Nahrmann Y, Kosinsky R L, Johnsen S A, Lecuit M, Höfer T, Rodewald HR
Pei W, Shang F, Wang X, Fanti AK, Greco A, Busch K, Klapproth K, Zhang Q, Quedenau C, Sauer S, Feyerabend T, Höfer T, Rodewald HR
Plum T, Wang X, Rettel M, Krijgsveld J, Feyerabend T, Rodewald HR
Pei W, Feyerabend TB, Rössler J, Wang X, Postrach D, Busch K, Rode I, Klapproth K, Dietlein N, Quedenau C, Chen W, Sauer S, Wolf S, Höfer T, Rodewald HR
Busch K, Klapproth K, Barile M, Flossdorf M, Holland-Letz T, Schlenner SM, Reth M, Höfer T, Rodewald HR
Gomez Perdiguero E, Klapproth K, Schulz C, Busch K, Azzoni E, Crozet L, Garner H, Trouillet C, de Bruijn MF, Geissmann F, Rodewald HR
Martins VC, Busch K, Juraeva D, Blum C, Ludwig C, Rasche V, Lasitschka F, Mastitsky SE, Brors B, Hielscher T, Fehling HJ, Rodewald HR
Get in touch with us

Prof. Dr. Hans-Reimer Rodewald
