Cancer Epigenomics
- Functional and Structural Genomics

Prof. Dr. Christoph Plass
Aberrant DNA methylation is an early event in tumorigenesis and a major contributor in the development of solid tumors as well as leukemias. As an epigenetic alteration, DNA methylation does not change the sequence of a gene and thus offers the exciting possibility for therapeutic removal of the methylation group by demethylating drugs.

Our Research
Deregulation of mechanisms that control the establishment of normal DNA methylation patterns leads to both extensive aberrant hypo- and hypermethylation and has been described for several human malignancies. Global DNA hypomethylation in human cancers was one of the earliest changes associated with tumor progression. Our group has shown that human malignancies are characterized by extensive promoter CpG island methylation with non-random and tumor-type specific patterns. DNA methylation changes co-occur with other epigenetic alterations such as nucleosome positioning and histone tail modifications. It is to the greatest extent unknown how tumors acquire aberrant DNA methylation patterns, however many of the enzymes that regulate epigenetic processes are mutated in human malignancies. Our Division is interested in the molecular mechanisms underlying the initiation and progression of malignant cell growth. In particular, we are focusing our attention on the contribution of epigenetic alterations in this process and to determine how epigenetic and genetic alterations cooperate during tumorigenesis. In our studies we are utilizing current state-of-the-art high throughput epigenomic assays (e.g. Illumina methylation arrays, next-generation whole genome bisulfite sequencing on minute amounts of cells, sc-multiome sequencing, antibody-guided chromatin tagmentation (ACT-seq)) on clinical samples, cell culture and rodent tumor models.
Future Outlook
Epigenetics is a quickly evolving field with links to many research directions in cancer research. A challenge here will be to integrate epigenetic questions with other data sets. For example in the past, the profiling of cancer genomes relied heavily on the description of genetic alterations. Now, epigenetic datasets will need to be included in order to completely understand the molecular defects in cancer. Furthermore, cellular identity is achieved through epigenetic regulation, and we will need to better define the epigenomes of cell types from which cancers arise since these preexisting epigenomes will influence the progression and aggressiveness of cancers as well as influence the therapeutic outcome. Our Division will focus on six major research directions:
- Evaluation of genome-wide epigenetic patterns in tumor genomes and the cell of origin from which individual cancers arise
- Identification of novel cancer genes and pathways targeted by epigenetic alterations
- Define the molecular mechanisms that lead to altered epigenetic patterns in cancer
- Determining the role of epigenetic pattern dynamics in differentiation of hematopoietic stem cells and other cell types
- Evaluating the role of epigenetics in the regulation of damage response
- Developing bioinformatical tools for the integrative analysis of epigenetic data
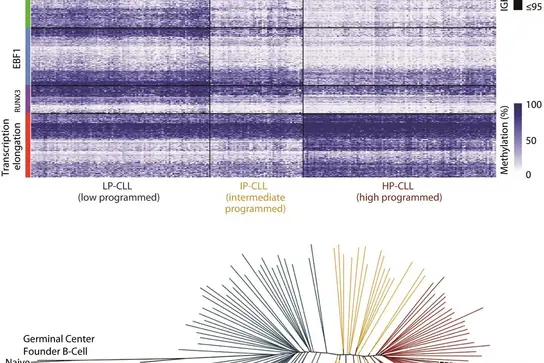
DNA methylation dynamics during B cell maturation underlie a continuum of disease phenotypes in chronic lymphocytic leukemia. Malignant cells typically expand clonally and normal tissues retain complex mixtures of various cell types, reported cancer-specific events are potentially inaccurate. Using whole genome bisulfite sequencing and other methods, we assess the dynamic DNA methylation events that occur as part of a broad epigenetic program established during the maturation of B cells. In comparison to malignant B cells from 268 chronic lymphocytic leukemia (CLL) patients, we reveal that tumors have the potential to derive from a continuum of possible maturation states that are reflected in the maturation stages of normal cells. The degree of maturation achieved in tumors closely associates with the acquisition of a more indolent pattern of gene expression and increasingly favorable clinical outcomes. (Oakes CC et al. Nature Genetics, 2016; 48:253-264)
Team
30 Employees
-

Prof. Dr. Christoph Plass
-
Marion Bähr
-
Dr. Ali Bakr
-
Kaushani Banerjee
-

Dr. Fiona Brown
Postdoctoral Researcher
-
Hossam Mohammed Hilmi Eldesouky
-
Elena Everatt
-
Dr. Clarissa Gerhäuser
-
Dr. Ashish Goyal
-
Susanna Grenner
-
Jessica Heilmann
-
Laura Herrfurth
-
Luc Husemann
-
Katherine Kelly
-
Daria Kiriy
-
Dr. Maria Llamazares Prada
-
Dr. Pavlo Lutsik
-
Sergio Manzano Sanchez
-
Nana Mensah
-
Oliver Mücke
-
Thekli Paschali
-
Antonella Sarnataro
-
Dr. Michael Scherer
-
Kathleen Schlüter
-
Etienne Sollier
-
Jennifer Ureta
-
Peter Waas
-
Dr. habil. Dieter Weichenhan
-
Joachim Weischenfeldt
-
Nan Zhang
Selected Publications
Progressive epigenetic programming during B cell maturation yields a continuum of disease phenotypes in chronic lymphocytic leukemia
Oakes CC, Seifert M, Assenov Y, Gu L, Przekopowitz M, Ruppert AS, Wang Q, Serva A, Koser S, Brocks D, Lipka D, Bogatyrova O, Mertens D, Zapatka M, Lichter P, Döhner H, Küppers R, Zenz T, Stilgenbauer S, Byrd JC and Plass C
Molecular evolution of early-onset prostate cancer identifies molecular risk markers and clinical trajectories
Gerhauser C, Favero F, Risch T, Simon R, Feuerbach L, Assenov Y, Heckmann D, Sidiropoulos N, Waszak SM, Hübschmann D, Urbanucci A, Girma EG, Kuryshev V, Klimczak LJ, Saini N, Stütz AM, Weichenhan D, Böttcher LM, Toth R, Hendriksen JD, Koop C, Lutsik P, Matzk S, Warnatz HJ, Amstislavskiy V, Feuerstein C, Raeder B, Bogatyrova O, Schmitz EM, Hube-Magg C, Kluth M, Huland H, Graefen M, Lawerenz C, Henry GH, Yamaguchi TN, Malewska A, Meiners J, Schilling D, Reisinger E, Eils R, Schlesner M, Strand DW, Bristow RG, Boutros PC, von Kalle C, Gordenin D, Sültmann H, Brors B, Sauter G, Plass C, Yaspo ML, Korbel JO, Schlomm T, Weischenfeldt J
Epigenetic reprogramming of airway macrophages promotes polarization and inflammation in muco-obstructive lung disease
Hey J, Paulsen M, Toth R, Weichenhan D, Butz S, Schatterny J, Liebers R, Lutsik P, Plass C, Mall MA
DNMT and HDAC inhibition induces immunogenic neoantigens from human endogenous retroviral element-derived transcripts
Goyal A, Bauer J, Hey J, Papageorgiou DN, Stepanova E, Daskalakis M, Scheid J, Dubbelaar M, Klimovich B, Schwarz D, Märklin M, Roerden M, Lin Y, Ma T, Mücke O, Rammensee H, Lübbert M, Loayza-Puch F, Krijgsveld J, Walz JS, Plass C
Get in touch with us

