Proteomics
Dr. Dominic Helm
Head of Facility
The Proteomics Core Facility at the DKFZ offers services related to protein analysis for internal groups and their cooperation partners. From the digestion of the proteins to the measurement on our LC-MS instruments to the data analysis, we find out how the proteome of your samples is composed.

Services
We offer the following sample preparation services:
- Protocols for extraction buffers and preparation of ready-to-use buffers
- Provision of various devices for protein extraction (see Instrumentation)
- Bead-based protein digestion (SP3)
- SDS-PAGE, Coomassie staining and in-gel digestion
- Peptide purifications
Source SP3: Hughes, Christopher S., et al. "Single-pot, solid-phase-enhanced sample preparation for proteomics experiments." Nature protocols 14.1 (2019): 68-85.
Source In-gel digestion: Shevchenko, Andrej, et al. "In-gel digestion for mass spectrometric characterization of proteins and proteomes." Nature protocols 1.6 (2006): 2856-2860.
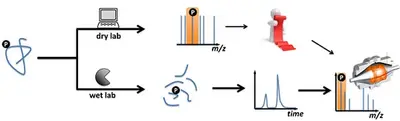
If you are interested in analyzing post-translationally modified proteins (PTM), we offer the option of either chemically enriching them prior to measurement (wet lab) or specifically searching for the corresponding modification during data analysis (dry lab).
Phosphorylation: In our lab we chemically enrich phosphorylated peptides after protein digestion.
Further tests for the chemical enrichment of other PTMs are being planned or developed.
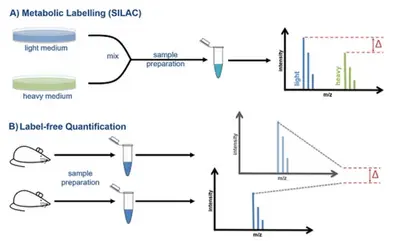
Since intensity and concentration do not correlate directly with each other, but are influenced by differences between the individual runs and the chemical properties of the peptides, special workflows are required to perform accurate quantification using mass spectrometry. We offer you the choice between two different quantitative methods: A) Metabolic labeling such as SILAC, B) Label-free quantification.
This highly specialised workflow is being supported and conducted in collaboration with our colleagues from the AG Riemer.
Please click here for more information.

Data-Dependent Acquisition (DDA)
In DDA precursor ions are selected dynamically for fragmentation based on their intensity. The instrument cycles through the most abundant ions in each MS scan, generating high-quality MS/MS spectra for peptide identification and PTM analysis.
Data-Independent Acquisition (DIA)
DIA systematically fragments all precursors within predefined m/z windows, ensuring comprehensive and unbiased data collection. This approach provides reproducible and quantitative accurate datasets with a deeper proteome coverage compared to DDA. Making it the ideal method for analyzing highly complex samples even in large scale studies.
Image source: Hu, Alex, William S. Noble, and Alejandro Wolf-Yadlin. "Technical advances in proteomics: new developments in data-independent acquisition." F1000Research 5 (2016).
We offer project tailored data base searches with state of the art software solutions.
Upon request we also perform customized statistical analysis, including pathway analysis and visualization.
Our workflow
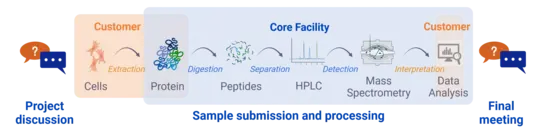
We assist you in planning your proteomics project, give advice on protein extraction methods and agree with you on the best analysis workflow for your needs.
After you have completed the extraction of the proteins in your laboratory and labeled the samples in randomized order, you can create a project in our booking system and submit the samples to us. Please note that we are only allowed to accept sample material of security level 1.
We will digest and measure the proteins using the agreed methods and then analyze the data.
We conduct a final meeting to evaluate the complete workflow of your project.
More detailed information on our protein lysate requirements and sample submission can be found in the Links section, PCF User Guide.
Please feel free to contact us.
Instrumentation

Our LC-MS systems are equipped with Orbitrap-based mass spectrometers, the Orbitrap Exploris 480 from Thermo Fisher Scientific. These instruments are characterized by high sensitivity and scanning speed in combination with mass accuracy in the ppm range and high resolution and are therefore perfectly suited for high-throughput proteome analysis with optimal data quality.
We are eagerly awaiting the arrival of our new Orbitrap Astral mass spectrometers in 2025.

The peptides to be analyzed in our mass spectrometers are separated online by polarity prior to measurement using Ultimate-3000 nano-flow instruments or Vanquish Neo UHPLC systems from Thermo Fisher Scientific. This enables in-depth analysis of complex samples in single LC-MS runs.
The Ultimate 3000 units in an analytical flow setup can be used for prior offline fractionation of peptides if desired.

For our SP3 digestion we use the BRAVO Liquid Handling Platform from Agilent Technologies. The device allows us to process up to 96 samples simultaneously.

We provide our customers with the following devices for sample preparation (Assisted-Access):
- Qiagen Tissue Lyser II
- Diagenode Bioruptor Pico
There is a booking plan for the Bioruptor in PPMS. Under "Links" you will find detailed instructions on how to access the device.
Technology Development
- Testing chemical enrichment methods for other relevant PTMs
- Improving protein extraction together with our customers
- Spatial proteomics
- Adaptation and development of various MS measurement methods
- Plasmaproteomics
Team
-
Dr. Dominic Helm
Head of Facility
-
Frederic Bethke
Technician (LC-MS)
-
Gerhard Erben
Engineer (LC-MS)
-
Sabine Fiedler
Technician (Sample Preparation)
-

Dr. Dario Frey
Postdoctoral Researcher
-
Ramona Irsevic
Technician (Sample Preparation)
-

Denise Jansen
PhD Student
-
Anna Neuerburg
Technician (Sample Preparation)
-
Martin Schneider
Scientist (Bioinformatician)
-

Dr. Piotr Zadora
Postdoctoral Researcher
Publications
Philipp M Roessner, Isabelle Seufert, Vicente Chapaprieta, Ruparoshni Jayabalan, Hannah Briesch, Ramon Massoni-Badosa, Pavle Boskovic, Julian Benckendorff, Tobias Roider, Lavinia Arseni, Mariana Coelho, Supriya Chakraborty, Alicia M Vaca, Mariela Sivina, Markus Muckenhuber, Sonia Rodriguez-Rodriguez, Alice Bonato, Sophie A Herbst, Marc Zapatka, Clare Sun, Helene Kretzmer, Thomas Naake, Peter-Martin Bruch, Felix Czernilofsky, Elisa Ten Hacken, Martin Schneider, Dominic Helm, Deyan Y Yosifov, Joseph Kauer, Alexey V Danilov, Moritz Bewarder, Kristina Heyne, Christof Schneider, Stephan Stilgenbauer, Adrian Wiestner, Jan-Philipp Mallm, Jan A Burger, Dimitar G Efremov, Peter Lichter, Sascha Dietrich, José I Martin-Subero, Karsten Rippe, Martina Seiffert
Silvia Schäfer, Lola Schmitz-Hübsch, Mehak Nihal Shaikh, Silke Brilloff, Elahe Rahimian, Vida Kufrin, Sandra Küchler, Maria Fedorova, Dominic Helm, Claudia R Ball, Martin Bornhäuser, Hanno Glimm, Meritxell Alberich Jorda, Marius Bill, Alexander A Wurm
Cristina Chiva, Roger Olivella, An Staes, Teresa Mendes Maia, Christian Panse, Karel Stejskal, Thibaut Douché, Bérangère Lombard, Andrea Schuhmann, Damarys Loew, Karl Mechtler, Mariette Matondo, Mandy Rettel, Dominic Helm, Francis Impens, Simon Devos, Anna Shevchenko, Paolo Nanni, Eduard Sabidó
Wai-Ling Macrina Lam, Gisela Gabernet, Tanja Poth, Melanie Sator-Schmitt, Morgana Barroso Oquendo, Bettina Kast, Sabrina Lohr, Aurora de Ponti, Lena Weiß, Martin Schneider, Dominic Helm, Karin Müller-Decker, Peter Schirmacher, Mathias Heikenwälder, Ursula Klingmüller, Doris Schneller, Fabian Geisler, Sven Nahnsen, Peter Angel
Emma L Phillips, Sizèd van Enk, Sara Kildgaard, Silja Schlue, Mona Göttmann, Victoria Jennings, Frederic Bethke, Gabriele Müller, Christel Herold-Mende, Daniel Pastor-Flores, Martin Schneider, Dominic Helm, Thomas Ostenfeld Larsen, Violaine Goidts
Hana Nůsková, Fabiola Garcia Cortizo, Lena Sophie Schwenker, Timo Sachsenheimer, Egor E Diakonov, Marcel Tiebe, Martin Schneider, Jasmin Lohbeck, Carissa Reid, Annette Kopp-Schneider, Dominic Helm, Britta Brügger, Aubry K Miller, Aurelio A Teleman
Tolga Lokumcu, Murat Iskar, Martin Schneider, Dominic Helm, Glynis Klinke, Lisa Schlicker, Frederic Bethke, Gabriele Müller, Karsten Richter, Gernot Poschet, Emma Phillips, Violaine Goidts
Changwen Wang, Meng Sun, Chunxuan Shao, Lisa Schlicker, Yue Zhuo, Yassin Harim, Tianping Peng, Weili Tian, Nadja Stöffler, Martin Schneider, Dominic Helm, Youjun Chu, Beibei Fu, Xiaoliang Jin, Jan-Philipp Mallm, Moritz Mall, Yonghe Wu, Almut Schulze, Hai-Kun Liu
Donato Inverso, Jingjing Shi, Ki Hong Lee, Moritz Jakab, Shani Ben-Moshe, Shubhada R Kulkarni, Martin Schneider, Guanxiong Wang, Marziyeh Komeili, Paula Argos Vélez, Maria Riedel, Carleen Spegg, Thomas Ruppert, Christine Schaeffer-Reiss, Dominic Helm, Indrabahadur Singh, Michael Boutros, Sudhakar Chintharlapalli, Mathias Heikenwalder, Shalev Itzkovitz, Hellmut G Augustin
Kuailu Lin, Franziska Baenke, Xixi Lai, Martin Schneider, Dominic Helm, Heike Polster, Venkatesh S Rao, Nicole Ganig, Fang Cheng Wong, Lena Seifert, Adrian M Seifert, Beatrix Jahnke, Nicole Kretschmann, Tjalf Ziemssen, Fee Klupp, Thomas Schmidt, Martin Schneider, Yi Han, Tim F Weber, Verena Plodeck, Heiner Nebelung, Nathalie Schmitt, Felix Korell, Bruno C Köhler, Carina Riediger, Jürgen Weitz, Nuh N Rahbari, Christoph Kahlert
Edoardo Ratto, S Roy Chowdhury, Nora S Siefert, Martin Schneider, Marten Wittmann, Dominic Helm, Wilhelm Palm
Catarina Pechincha, Sven Groessl, Robert Kalis, Melanie de Almeida, Andrea Zanotti, Marten Wittmann, Martin Schneider, Rafael P de Campos, Sarah Rieser, Marlene Brandstetter, Alexander Schleiffer, Karin Müller-Decker, Dominic Helm, Sabrina Jabs, David Haselbach, Marius K Lemberg, Johannes Zuber, Wilhelm Palm
Michael Bonadonna, Sandro Altamura, Elisabeth Tybl, Gael Palais, Maria Qatato, Maria Polycarpou-Schwarz, Martin Schneider, Christina Kalk, Wibke Rüdiger, Alina Ertl, Natasha Anstee, Ruzhica Bogeska, Dominic Helm, Michael D Milsom, Bruno Galy
Links
Get in touch with us

